FIG. 4.
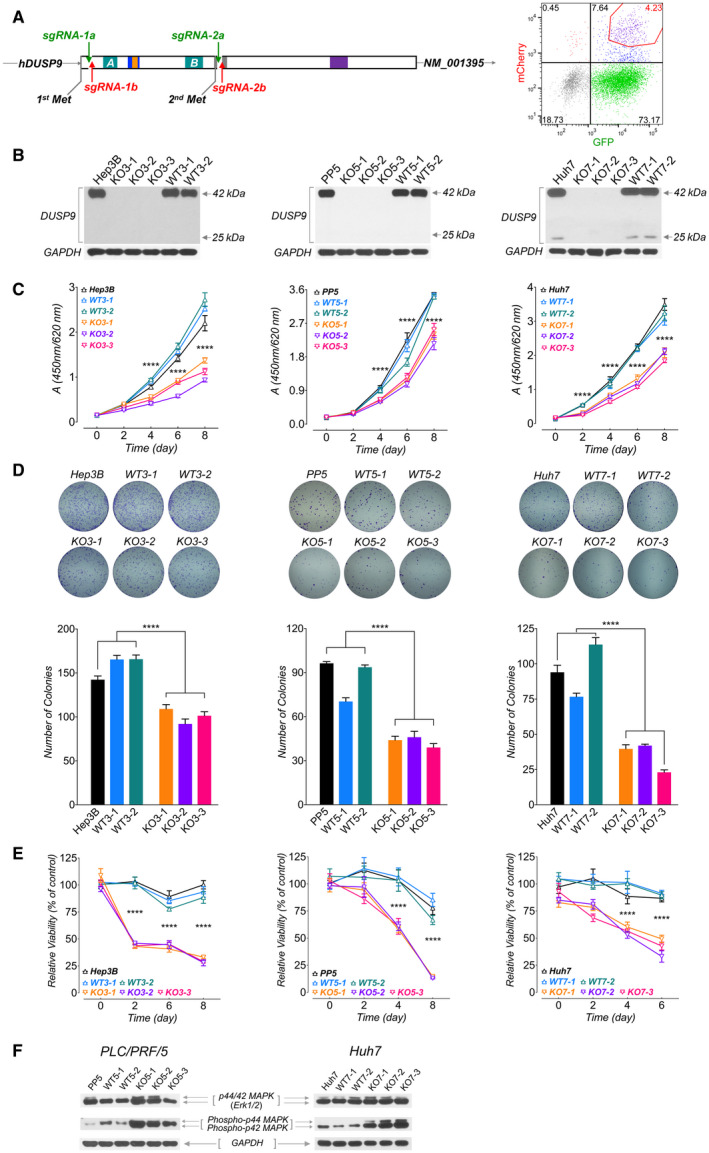
Characterization of DUSP9 KO HCC cells. (A) Targeting strategy for CRISPR‐mediated DUSP9 KO. Cas9n‐mediated DNA cleavage sites were designed after the first start codon (Hep3B and PP5 cells, using sgRNA‐1a/1b) or after the second start codon (HepG2 and Huh7 cells, using sgRNA‐2a/2b). FACS plot (right) shows gating of transfected cells with dual fluorescent tags (GFP for sgRNA‐1a or 2a, mCherry for sgRNA‐1b or 2b) for further screening. (B) Western blots for DUSP9 in WT and KO cell lines. (C) WST‐1 assay on DUSP9 WT and KO cells. Data represent mean of six replicates per group; ****P < 0.0001. (D) Colony formation assays on DUSP9 KO and WT cells. Data show mean + SEM of three replicates per group; ****P < 0.0001. (E) Relative viability assays of doxorubicin‐treated DUSP9 KO versus WT Hep3B (200 ng/mL), PP5 (400 ng/mL), and Huh7 (1 µg/mL) cells. Data show mean + SEM of six replicates per group; ****P < 0.0001. (F) Western blot of DUSP9 KO and WT cells for Erk1/2 (p44/42 MAPK) and phosphorylated Erk1/2 (phospho‐p44/42 MAPK). Abbreviation: FACS, fluorescence‐activated cell sorting.
