Fig. 4. NSD1/2 and LEDGF/HDGF2 coregulate protumorigenic pathways and can be disrupted by a H3K36me2-mimicking peptide.
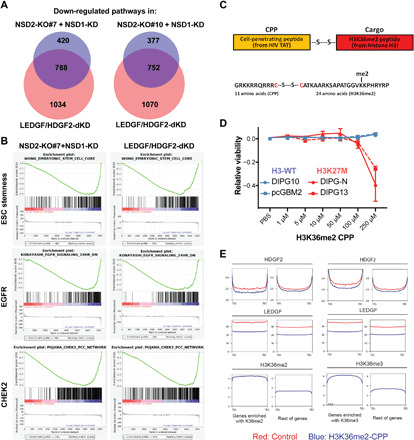
(A) Venn diagrams showing overlaps of gene signatures/pathways down-regulated in LEDGF/HDGF2 dKD DIPG13 cells and NSD2-KO#7 + siNSD1 or NSD2-KO#10 + siNSD1 DIPG13 cells (P = 4.2 × 10−9 and 1.8 × 10−4, respectively). The alteration of gene signatures/pathways was detected by GSEA. P values were obtained by the hypergeometric distribution to compute the significance of the overlap of two pathway sets. (B) Representative images of highly ranked GSEA signatures/pathways detected in (A), including a CHEK2 pathway, an embryonic stem cell (ESC) steamness signature, and a set of EGFR signaling target genes. (C) Top: A schematic illustration of the design of CPP. A HIV-based cell entry peptide was linked to a H3K36me2 peptide (histone H3 21 to 43 amino acids, cargo peptide) by a disulfide bond. Bottom: Amino acid sequence of H3K36me2-linked CPP. (D) A CellTiter-Glo cell survival assay for H3-WT (DIPG10 and pcGBM2) and H3K27M (DIPG-N and DIPG13) cells treated with control (vehicle only) or H3K36me2-CPP. Cells were assayed at 72 hours after dosing with a titration of control or H3K36me2-CPP and data were presented by ratios of CellTiter-Glo signals in control versus H3K36me2-CPP treated cells. PBS, phosphate-buffered saline. (E) Metaprofiles of ChIP-seq analysis for changes in LEDGF and HDGF2 occupancy of genes enriched with H3K36me2 or H3K36me3 and the rest of genes in DIPG13 cells treated with a control vehicle (red) or a H3K36me2-CPP (blue).
