Abstract
Vascular calcification (VC) predicts cardiovascular morbidity and mortality in chronic kidney disease (CKD). To date, the underlying mechanisms remain unclear. We detected leukocyte DNA N6-methyladenine (6mA) levels in patients with CKD with or without aortic arch calcification. We used arteries from CKD mice infected with vascular smooth muscle cell–targeted (VSMC-targeted) adeno-associated virus encoding alkB homolog 1 (Alkbh1) gene or Alkbh1 shRNA to evaluate features of calcification. We identified that leukocyte 6mA levels were significantly reduced as the severity of VC increased in patients with CKD. Decreased 6mA demethylation resulted from the upregulation of ALKBH1. Here, ALKBH1 overexpression aggravated whereas its depletion blunted VC progression and osteogenic reprogramming in vivo and in vitro. Mechanistically, ALKBH1-demethylated DNA 6mA modification could facilitate the binding of octamer-binding transcription factor 4 (Oct4) to bone morphogenetic protein 2 (BMP2) promoter and activate BMP2 transcription. This resulted in osteogenic reprogramming of VSMCs and subsequent VC progression. Either BMP2 or Oct4 depletion alleviated the procalcifying effects of ALKBH1. This suggests that targeting ALKBH1 might be a therapeutic method to reduce the burden of VC in CKD.
Keywords: Cardiology, Vascular Biology
Keywords: Cardiovascular disease, Chronic kidney disease, Epigenetics
Introduction
The leading cause of death in patients with chronic kidney disease (CKD) has been traced to cardiovascular complications (1). Medial vascular calcification (VC) is prevalent in CKD and contributes to subsequent cardiovascular morbidity and mortality (2). VC progressively increases with advanced stages of CKD and is linked with hyperphosphatemia (3). Hyperphosphatemia results in ectopic deposition of calcium-phosphate crystals in the vasculature, thus demonstrating that phosphate is a pivotal contributor to VC development. VC results in vessel wall stiffening and impaired elastic recoil (4). Consequently, the hemodynamic instability increases cardiac afterload and results in irreversible outcomes (5).
Multiple regions of the aorta are prone to calcification during CKD. Indeed, the aortic arch is more prone to calcification throughout the entire aorta at the early stage. Aortic arch calcification can easily be detected via chest computed tomography, which accurately represents the magnitude of VC (6). Aortic arch calcification is a prognostic indicator of worse cardiovascular outcomes (7). Various strategies have been devised to slow the progression of this burgeoning health concern. However, no substantial early diagnosis and practical pharmacological approach have been proved to retard VC.
Vascular smooth muscle cells (VSMCs) within the artery undergo osteogenic reprogramming to potentiate VC. This active process is recognized as similar to physiologic bone mineralization (8). Hyperphosphatemia drives VSMCs to convert from the contractile phenotype to the osteogenic phenotype, which is characterized by loss of VSMCs markers (smooth muscle 22 alpha [SM22α], alpha-smooth muscle actin [α-SMA], and Calponin1), accompanied with increased expression of osteogenic markers (osteopontin [OPN], osteocalcin [OCN], and Collagen I) (9). The bone morphogenetic proteins (BMPs) are members of the transforming growth factor beta family that engaged in embryogenesis, organogenesis, and osteoblast differentiation (10). Bone morphogenetic protein 2 (BMP2), a member of the BMPs family, is a potent osteogenic protein, which has been shown to promote VSMCs osteogenic reprogramming and VC. BMP2 accomplishes this by elevating the expression of runt-related transcription factor 2 (RUNX2), the decisive transcription factor for bone formation (11).
Currently, there is limited understanding of how and why osteogenic reprogramming occurs. Epigenetic alterations to DNA methylation (12), histone modification (13), and microRNA have emerged as critical regulators of this process (14). DNA methylation abnormalities have been well described in patients with CKD (12). Previous studies substantiated that elevated phosphate environment increased DNA methyltransferase activity and methylation of transgelin (TAGLN) promoter region, thus leading to decreased expression of SM22α and a consequent increase in VC (15).
Recent developments in deep sequencing technology allowed the detection of a novel DNA adenine methylation (N6-methyladenine [6mA]) in the human genome (16). This modification is mediated by the methyltransferase N6 adenine–specific DNA methyltransferase 1 (N6AMT1) and demethylase alkB homolog 1 (ALKBH1) (17). Several studies revealed that there is a robust relationship between ALKBH1-demethylated DNA 6mA modification and cardiovascular diseases, such as hypertension and atherosclerosis (18, 19). As previously reported, ALKBH1-mediated 6mA demethylation is required for the osteogenic differentiation of human mesenchymal stem cells during bone metabolism (20). However, the role of 6mA in VC has not been explored.
Considering the overlapping bone-vascular axis in VC, it is tempting to postulate that ALKBH1 may have a regulatory effect on the progression of VC. Therefore, this study was conducted to elucidate the role of ALKBH1-demethylated DNA 6mA modification in hyperphosphatemia-induced VC during CKD. We aim to provide new insights into early diagnosis and potential targets for therapeutic medication towards VC in CKD.
Results
Reduced leukocyte DNA 6mA is correlated with the severity of vascular calcification in the clinical setting.
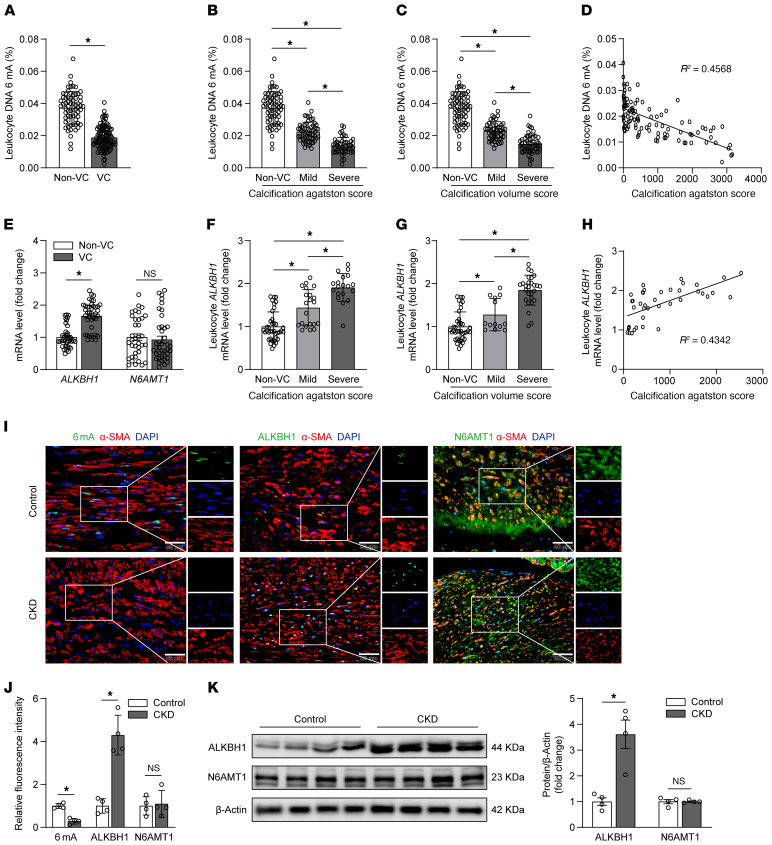
CKD-induced hyperphosphatemia can elicit multiple alterations in cellular epigenetic regulation (21, 22). Leukocyte DNA 6mA levels were evaluated in patients with CKD, with or without aortic arch calcification, to elucidate modification alterations during VC. The CKD patient characteristics in the non-VC and VC groups are shown in Table 1. To bolster confidence in the CKD cohort, we introduced the soluble urokinase plasminogen activator receptor (suPAR), which was proved as a practical and strong clinical indicator for renal disease (23–27). The plasma phosphate levels of patients with CKD showed mild difference between 2 groups. This frustrating result may be accounted by the therapeutic controlling of hyperphosphatemia in recruited patients with CKD. In the non-VC group of patients with CKD, 32% accepted maintenance dialysis, whereas the percentage in the VC group is 68% (data not shown). The chest computed tomographic scan of patients with CKD delineated the dense patchy calcification present in the wall of the aortic arch (Figure 1, A–D). Compared with noncalcification controls, 6mA levels were significantly reduced in patients with CKD with calcification by approximately 49.6% (mean 0.0373% ± 0.01% vs. 0.0188% ± 0.007%) (Figure 2A). Next, we quantified aortic arch calcification in patients with CKD by area and volume, which were presented as Agatston score and Volume score, respectively. As the severity of VC increased, 6mA levels progressively decreased. Individuals exhibiting the largest or thickest calcified lesions had the lowest 6mA levels (Figure 2, B and C). A negative correlation was observed between the DNA 6mA level and the calcification score in patients with CKD with VC (R2 = 0.4568 and R2 = 0.4637, Pearson’s correlation coefficient analysis) (Figure 2D and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI146985DS1). Taken together, these data demonstrated that leukocyte 6mA levels are associated with VC progression in the clinical setting.
Table 1. Basal characteristics in patients with CKD with or without calcification.
Figure 1. Representative images of multi-detector computed tomography (MDCT) scan of CKD patients with or without calcification.
Representative axial image (A) and sagittal reconstruction (B) of contrast-enhanced CT scan in CKD patients without calcification. Representative axial image (C) and sagittal reconstruction (D) of contrast-enhanced CT scan showing aortic arch calcification in CKD patients.
Figure 2. ALKBH1-demethylated DNA 6mA modification is reduced in human vascular calcifying progression during CKD.
(A) Leukocyte DNA 6mA level in patients with CKD with (VC, n = 106) or without (non-VC, n = 67) aortic arch calcification. (B and C) Leukocyte DNA 6mA level in subgroups defined by calcification Agatston score (B, n = 67 for non-VC; n = 61 for mild; n = 45 for severe) and Volume score (C, n = 67 for non-VC; n = 53 for mild; n = 53 for severe). (D) Correlation between leukocyte DNA 6mA level and calcification Agatston score from patients with CKD with aortic arch calcification (n = 106). (E) Quantitative real-time PCR analysis of ALKBH1 and N6AMT1 mRNA expression in leukocytes from patients with CKD with (VC, n = 40) or without (non-VC, n = 38) aortic arch calcification. (F and G) Leukocyte ALKBH1 mRNA expression level in subgroups defined by calcification Agatston score (F, n = 38 for non-VC; n = 21 for mild; n = 19 for severe) and Volume score (G, n = 38 for non-VC; n = 13 for mild; n = 27 for severe). (H) Scatterdot plot of correlation between leukocyte ALKBH1 mRNA expression level and calcification Agatston score from patients with CKD with aortic arch calcification (n = 40). (I–K) Representative immunofluorescence pictures (I) and quantification (J) and Western blot analysis (K) of ALKBH1, N6AMT1, and 6mA in radial arteries from CKD (n = 4) and control (n = 4) groups. Scale bars: 50 μm. Statistical significance was assessed using 2-tailed t tests (A, E, J, and K), 1-way ANOVA followed by Bonferroni’s test (B, C, F, and G), and Pearson’s correlation coefficient analysis (D and H). All values are presented as mean ± SD. *P < 0.05.
ALKBH1 is upregulated during vascular calcification.
The mRNA expression of ALKBH1, the 6mA demethylase, was significantly elevated and expression of N6AMT1, the 6mA methyltransferase, was slightly decreased in patients with CKD with VC. Thus, alteration to the 6mA profile in leukocytes is mainly due to ALKBH1 (Figure 2E). Interestingly, leukocyte ALKBH1 mRNA expression was increased as VC progressed (Figure 2, F and G) and positively associated with the calcification score of CKD patients with VC (Figure 2H and Supplemental Figure 1B). Next, ALKBH1, N6AMT1, and 6mA were detected in radial artery specimens from patients with CKD who underwent arterial-venous fistular surgery with diagnosed aortic arch calcification. Compared with normal controls, CKD specimens exhibited increased expression of ALKBH1 and decreased 6mA modification (Figure 2, I–K). In addition, the CKD patients with VC showed advanced age according to the basal clinical characteristics of patients with CKD (Table 1). However, 6mA levels were barely related to age in patients with CKD with the presence or absence of VC (Supplemental Figure 1C).
To further verify the observations of 6mA levels in vivo, 2 mice models were used. The diet-induced CKD model was generated by feeding mice with 0.2% adenine diet for 8 weeks. The surgery-promoted CKD model was conducted by performing 5/6 nephrectomy on mice. The plasma phosphorus, suPAR, blood urea nitrogen, and creatinine were elevated in both models (Supplemental Tables 1–3). The renal histological staining of those mice showed similar increased inflammatory cell infiltration, tubule lumen expansion, tubular vacuoles, interstitial fibrosis, and mineral deposition in renal glomerulus, tubules and small vessels (Supplemental Figures 2 and 3). In addition, there is increased medial arterial calcification with a remarkable elevation of plasma alkaline phosphatase (ALP) (Supplemental Tables 1–3). Therefore, VC is successfully induced in the CKD mice models. Consistent with the clinical observations in patients with CKD with VC, the leukocyte DNA 6mA level was decreased compared with controls (Figure 3A and Supplemental Figure 4A). Consistently, the calcification lesion size in the medial arterial layer of CKD mice had a negative correlation with the lower leukocyte 6mA level (Figure 3, B and C, and Supplemental Figure 4B). In addition, the mRNA expression of Alkbh1 was significantly upregulated but not N6amt1 in murine leukocytes (Figure 3D). Immunohistochemistry staining (Figure 3E and Supplemental Figure 4, C and D) and Western blot analysis (Figure 3F and Supplemental Figure 4E) demonstrated elevated levels of ALKBH1 in aortic smooth muscle layers of CKD mice, and 6mA levels were markedly decreased. In comparison, N6AMT1 showed no alteration in expression. Of note, the enhanced ALKBH1 expression in the smooth muscle layer was inversely correlated to reduced leukocyte 6mA level (Supplemental Figure 5), suggesting the corresponding systematic change of 6mA modification in vivo. These observations were verified by ex vivo mice aortic rings cultured in osteogenic medium, which showed trends similar to the elevated ALKBH1 expression and corresponding decreased 6mA levels (Figure 3, G–I). Taken together, these data suggest that the ALKBH1 is upregulated in leukocytes and VSMCs during VC.
Figure 3. ALKBH1-demethylated DNA 6mA modification is reduced in murine calcified arteries.
(A) Leukocyte DNA 6mA level in mice fed with adenine diet (CKD, n = 13) or normal chow diet (control, n = 12) for 8 weeks. Leukocytes were isolated from peripheral blood. (B and C) Mice leukocyte DNA 6mA level in different subgroups defined by the percentage of calcification lesion size in aortic smooth muscle layer (B, n = 12 for non-VC; n = 6 for mild; n = 7 for severe). Scatter dot plot of correlation between mice leukocyte DNA 6 mA level and percentage of calcification lesion size in aortic smooth muscle layer from mice fed with adenine diet for 8 weeks (C, n = 13). (D) The mRNA expression levels of Alkbh1 and N6amt1 in leukocytes from mice with different diets (n = 12 per group). (E) Representative immunohistochemistry pictures and quantification of ALKBH1, N6AMT1, and 6mA in mice aortic smooth muscle layer (n = 10 for control; n = 12 for CKD). Scale bars: 50 μm. (F) Western blot analysis of ALKBH1 and N6AMT1 expression in mice aortic arch (n = 4 for control; n = 5 for CKD). (G–I) Calcium content (G), Western blot analysis of ALKBH1 and N6AMT1 (H), and DNA 6 mA level (I) in mice aortic rings incubated with osteogenic medium for the indicated time (0, 3, 5, 7, 10, and 14 days) (n = 4–6 per group). Statistical significance was assessed using 2-tailed t tests (A and D–F), 1-way ANOVA followed by Bonferroni’s test (B) or Dunnett’s test (G–I), and Pearson’s correlation coefficient analysis (C). All values are presented as mean ± SD. *P < 0.05 vs. Pi (0 day) in (G–I).
Depletion of ALKBH1 alleviates vascular calcification.
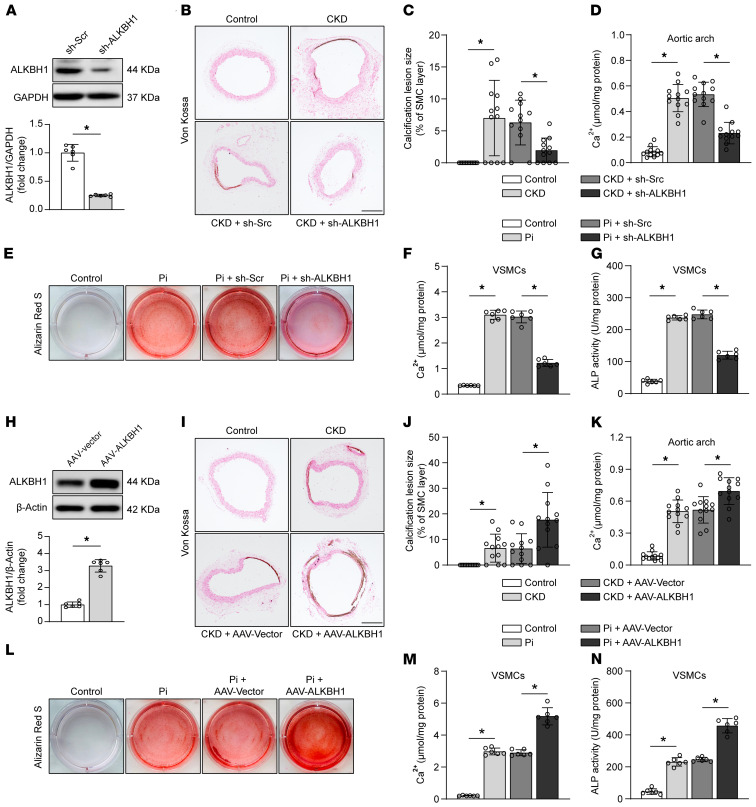
ALKBH1 can be upregulated during VC, so it is interesting to address whether ALKBH1 depletion might attenuate the VC progression. Most of the global Alkbh1 knockout mice died during the embryonic stage, and survivors exhibit developmental tissue defects (28). To elucidate ALKBH1 function, we used the adeno-associated virus (AAV) infection as a genomic manipulation model. AAV carrying scrambled shRNA or Alkbh1 shRNA (sh-ALKBH1) were inoculated via tail vein into 2 CKD mice models. Depletion of ALKBH1 in the aorta was confirmed via Western blot analysis (Figure 4A). Knockdown of ALKBH1 strikingly suppressed the calcium deposition and mineralization of the aorta compared with the scrambled controls (Figure 4, B–D, and Supplemental Figure 6), along with the plasma ALP (Supplemental Tables 1 and 3). To further confirm the in vivo results, we cultured primary murine VSMCs in an osteogenic medium for 2 weeks with sh-Scr or sh-ALKBH1. ALKBH1 knockdown cohorts exhibited significantly decreased calcium nodule formation as demonstrated by alizarin red staining (Figure 4E) and reduced calcium deposition, and the corresponding decreased ALP activity (Figure 4, F and G). Collectively, these results suggest that deficiency of ALKBH1 inhibits VC.
Figure 4. ALKBH1 is essential for the regulation of vascular calcification.
(A) Western blot analysis identifying the ALKBH1 deficiency in arteries (n = 6 per group). Mice were injected via tail vein with AAV carrying scrambled shRNA (sh-Scr) or Alkbh1 shRNA (sh-ALKBH1) at 4 weeks after adenine diet and then fed for another 4 weeks. (B–D) Von Kossa staining (B and C) and calcium content quantification of aortic arch (D) were performed in different experimental groups for detecting mineralization (n = 10–12 per group). Scale bar: 100 μm. (E) Photomicrographs of Alizarin red staining in mice primary VSMCs pretransfected with indicated treatment and exposed in osteogenic medium for another 14 days (n = 6 per group). (F and G) Bar graphs representative of calcium content (F) and ALP activity (G) in mice primary VSMCs from all of the experimental cohorts (n = 6 per group). (H) ALKBH1 overexpression in arteries confirmed by Western blot (n = 6 per group). Mice were injected with AAV-Vector or AAV-ALKBH1 at 4 weeks after the adenine diet and then fed for another 4 weeks. (I–K) Percentage of positive von Kossa staining (I and J) and calcium content (K) quantified in the aortic arch from the different cohorts (n = 10–12 per group). Scale bar: 100 μm. (L) Representative images of Alizarin red staining in mice primary VSMCs after indicated transfection and osteogenic medium exposure for another 14 days (n = 6 per group). (M and N) Scatter dot plots representative of calcium content (M) and ALP activity (N) in mice primary VSMCs from all of the experimental cohorts (n = 6 per group). Statistical significance was assessed using 2-tailed t tests (A and H) and 1-way ANOVA followed by Dunnett’s test (C–G, and J–N). All values are presented as mean ± SD. *P < 0.05.
Overexpression of ALKBH1 enhances vascular calcification.
To evaluate whether ALKBH1 could aggravate VC progression, we established an overexpression model by injecting AAV-vector or AAV-ALKBH1 to CKD mice induced by adenine diet or operation. The efficiency of transfection was verified, as indicated in Figure 4H. As expected, AAV-ALKBH1 markedly increase in vivo calcification, as determined by von Kossa staining, calcium assay (Figure 4, I–K, and Supplemental Figure 6), and quantification of plasma ALP activity (Supplemental Tables 2 and 3). In vitro results showed that AAV-vector infection did not alter the calcium deposition in cultured murine VSMCs. In contrast, ALKBH1 overexpression greatly exacerbated calcification progression (Figure 4, L–N), further suggesting that ALKBH1 promotes VC. The blood pressure of patients with CKD showed a difference between the non-VC and VC groups (Table 1). In addition, ALKBH1 was associated with hypertension (19). However, we found systolic blood pressure, diastolic blood pressure, and other parameters, including blood glucose, cholesterol, and triglyceride, were not affected by ALKBH1 deficiency or overexpression in vivo (Supplemental Tables 1–3, and Supplemental Figure 7).
ALKBH1 regulates osteogenic reprogramming of VSMCs.
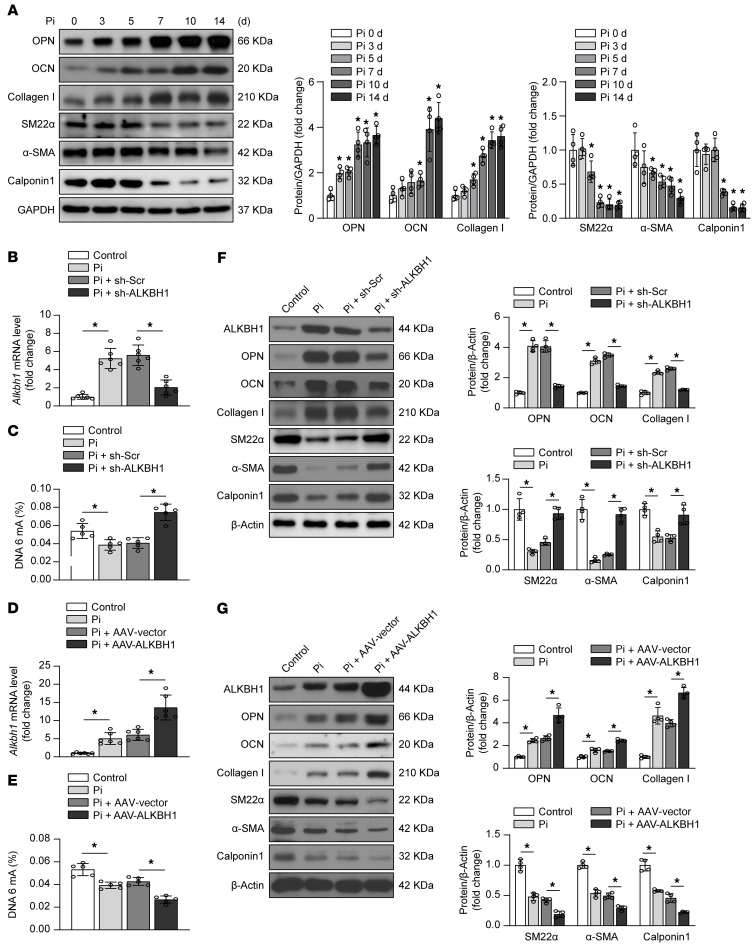
We next determined whether ALKBH1 could regulate the osteogenic reprogramming of VSMCs in vitro and in vivo. High inorganic phosphate (Pi) condition successfully induces VSMCs osteogenic reprogramming (Figure 5A). Deletion (Figure 5, B and C) or overexpression (Figure 5, D and E) of Alkbh1 in calcified mice primary VSMCs were confirmed by mRNA analysis and 6mA quantification. ALKBH1 deficiency dramatically suppressed the expression of osteogenic genes OPN, OCN, and Collagen I, and conversely increased expression of the contractile genes SM22α, α-SMA, and Calponin1 (Figure 5F). In contrast, the levels of osteogenic markers were upregulated whereas the contractile makers were decreased as a result of the overexpression of ALKBH1 (Figure 5G). The aortas from 5/6 nephrectomy–induced CKD mice further confirmed that OCN and SM22α expression were regulated by ALKBH1 (Supplemental Figure 8). Moreover, in HASMCs, ALKBH1 deficiency could attenuate the osteogenic reprogramming together with the calcium deposition (Supplemental Figure 9). Thus, these data show that ALKBH1 elevation promotes osteogenic reprogramming of VSMCs during calcification.
Figure 5. ALKBH1 regulates VSMCs osteogenic reprogramming.
(A) Western blot analysis of osteogenic phenotype marker (OPN, OCN, and Collagen I) and contractile phenotype marker (SM22α, α-SMA, and Calponin1) expression in mice primary VSMCs cultured in osteogenic medium for 14 days. (B) Quantitative real-time PCR analysis of Alkbh1 expression in mice primary VSMCs, which were pretransfected with AAV encoding scrambled or Alkbh1 shRNA for 48 hours, and then cultured in osteogenic medium for another 14 days. (C) Quantitative DNA 6mA level in ALKBH1-deficient mice primary VSMCs. (D) Quantitative real-time PCR analysis of Alkbh1 expression in mice primary VSMCs, which were preinfected with AAV-Vector or AAV-ALKBH1 for 48 hours, and then cultured in osteogenic medium for another 14 days. (E) Quantitative DNA 6mA level in ALKBH1-overexpressed mice primary VSMCs. (F) Western blot analysis of osteogenic phenotype marker and contractile phenotype marker expression in mice primary VSMCs with ALKBH1 depletion. (G) Western blot analysis of osteogenic phenotype marker and contractile phenotype marker expression in mice primary VSMCs with ALKBH1 overexpression. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s test. n = 4–6 for each group. All values are presented as mean ± SD. *P < 0.05 vs. Pi (0 day) in (A).
BMP2 mediates ALKBH1-regulated VSMCs osteogenic reprogramming.
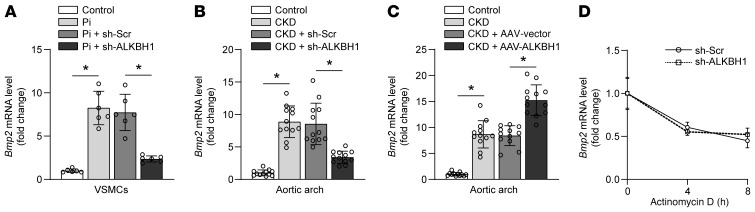
We detected transcription factors and signaling pathways that were vital during osteogenic reprogramming. ALKBH1 depletion altered BMP2 and RUNX2 expression but not SRY-box transcription factor 9 (SOX9) and distal-Less homeobox 5 (DLX5) (Figure 6A and Supplemental Figure 10). Immunofluorescence staining further confirmed that ALKBH1 depletion suppressed BMP2 in aortas from CKD mice, along with reciprocal changes in α-SMA (Figure 6, B and C). To determine whether BMP2 mediates the ALKBH1-regulated VSMCs osteogenic reprogramming, mice primary VSMCs and aortic rings were infected with AAV sh-BMP2 together with AAV-ALKBH1. BMP2 knockdown effectively antagonized the ALKBH1-enhanced osteogenic reprogramming (Figure 6D) and mineralized nodule formation (Figure 6E), accompanied by decreased ALP activity (Figure 6F) and calcium content (Figure 6G). It has been previously reported that BMP2 acts by inducing the expression of RUNX2 in VSMCs, thereby affecting osteogenic reprogramming (29). We observed that BMP2 depletion could decrease RUNX2 upregulation, which indicated that BMP2 is the upstream regulator. Further real-time PCR analysis revealed that Bmp2 levels were transcriptionally regulated in vitro (Figure 7A) and in vivo (Figure 7, B and C). In addition, ALKBH1 depletion had no effect on the stability of Bmp2 mRNA (Figure 7D). Above all, these data indicate that ALKBH1-induced calcification is directly dependent on BMP2.
Figure 6. BMP2 mediates the procalcifying effects of ALKBH1.
(A) Western blot analysis of ALKBH1, BMP2, RUNX2, SOX9, and DLX5 expression in calcified mice primary VSMCs with AAV sh-Scr or AAV sh-ALKBH1 transfection (n = 4 per group). (B and C) Representative immunofluorescence images (B) and quantification (C) of α-SMA and BMP2 costained in aortas from indicated experimental cohorts (n = 6 per group). Scale bar: 50 μm. (D) Western blot analysis of osteogenic phenotype marker (RUNX2, OPN, OCN, and Collagen I) expression in mice primary VSMCs, which preinfected with AAV sh-Scr or AAV sh-BMP2 together with AAV-Vector or AAV-ALKBH1 and then incubated in calcifying medium for another 14 days. (E and F) Alizarin red staining (E) and ALP activity assay (F) performed in all of the groups for detecting calcification formation (n = 4–5 per group). (G) Quantification of calcium content in mice aortic ring cultured in calcifying medium with indicated transfection (n = 5 per group). Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s test (A–C) or Bonferroni’s test (D–G). All values are presented as mean ± SD. *P < 0.05.
Figure 7. BMP2 is regulated by ALKBH1 in transcriptional level.
(A) Quantitative real-time PCR analysis of Bmp2 expression in primary mice VSMCs with ALKBH1 depletion (n = 6 per group). (B and C) Quantitative real-time PCR analysis of Bmp2 expression in the aortic arch from mice with ALKBH1 knockdown (B) or ALKBH1 overexpression (C) (n = 12 per group). (D) Quantitative real-time PCR analysis of Bmp2 expression in mice primary VSMCs treated with actinomycin D (5 mg/mL) for a different time after AAV sh-Scr or AAV sh-ALKBH1 transfection (n = 3 per group). Gene expression was normalized to Gapdh. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s test. All values are presented as mean ± SD. *P < 0.05.
6mA demethylation promotes octamer-binding transcription factor 4 (Oct4) binding to the BMP2 promoter and activates transcription.
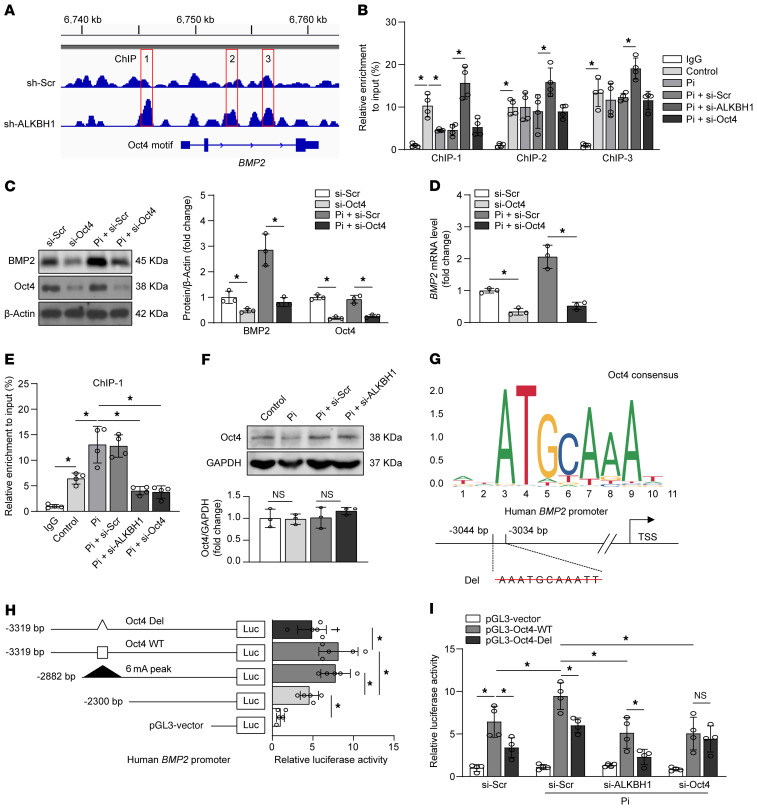
Next, we sought to elucidate the mechanism by which ALKBH1 depletion downregulates BMP2. Bioinformatics screening indicated 3 6mA peaks (marked as ChIP1, 2, and 3) that were induced by ALKBH1 knockdown in the human BMP2 gene (Figure 8A) (30). ChIP-qPCR confirmed that ALKBH1 silencing altered 6mA saturation in these 3 regions. However, only the first 6mA peak was subjected to osteogenic stimulation due to Pi-induced reduction of 6mA enrichment (Figure 8B). According to the literature, DNA methylation may regulate transcription by interfering with transcription factor binding (31). The sequence of the first region from –2882 bp to –2300 bp of the BMP2 promoter was analyzed. A binding site of Oct4, also known as POU domain class 5 transcription factor 1 (POU5F1), was predicted based on JASPAR with a defined 99% profile score threshold. We found that the application of OCT4 siRNA repressed BMP2 protein and mRNA expression in HASMCs (Figure 8, C and D). Furthermore, ChIP-qPCR with Oct4 pull-down further demonstrated that Pi-reduced 6mA modification facilitated Oct4 accumulation in the first region, whereas 6mA modification that was increased by ALKBH1 silencing abrogated this effect (Figure 8E). In other words, demethylation of the BMP2 promoter results in enhanced Oct4 binding. Interestingly, the Western blot results showed that neither ALKBH1 knockdown nor high Pi condition altered total Oct4 protein level (Figure 8F).
Figure 8. ALKBH1-dependent 6mA demethylation promotes Oct4 binding to the BMP2 promoter and activates BMP2 transcription.
(A) Integrative genomics viewer plots showing the increasing 6mA peaks (selected one marked as ChIP1-3) in human BMP2 gene (hg19) region with ALKBH1 knockdown via shRNA lentiviral constructs. (B) ChIP-qPCR assay displaying the 6mA enrichment on the 3 BMP2 fragments in treated HASMCs (n = 4 per group). (C and D) Quantitative Western blot (C) and real-time PCR analysis of BMP2 expression (D) in HASMCs with scramble or OCT4 siRNA transfection under calcifying condition (n = 3 per group). (E) ChIP-qPCR assay with Oct4 or IgG antibody for the ChIP-1 enrichment in treated HASMCs (n = 4 per group). (F) Western blot analysis of Oct4 in HASMCs incubated with osteogenic medium after transfection with scrambled siRNA (si-Scr) or ALKBH1 siRNA (si-ALKBH1). (G) Logos of the standard Oct4 motif and schematic of human BMP2 promoter showing wide-type (WT) and deleted (Del) binding sites for Oct4 within the first 6mA peak. (H) Bar graphs representative of the luciferase activity analyzed in HASMCs after cotransfection with control Renilla luciferase plasmid and serial deletion constructs of BMP2 promoter-driven luciferase reporters containing WT or Del Oct4 site (n = 5 per group). (I) Relative promoter activities measured by dual-luciferase reporter assay in HASMCs, which pretreated with indicated siRNA and then infected with pGL3-Oct4-WT or pGL3-Oct4-Del under calcifying conditions (n = 4 per group). Statistical significance was assessed using 1-way ANOVA followed by Bonferroni’s test. All values are presented as mean ± SD. *P < 0.05.
To detect whether Oct4 binding regulates BMP2 transcription, serial human BMP2 promoter-driven (–3319 bp) luciferase reporter assay was conducted with or without Oct4 binding site (Figure 8G). As shown in Figure 8H, Oct4 induced robust luciferase expression in pGL3-Oct4-WT, whereas mutation suppressed luciferase activity, suggesting that Oct4 binds directly to the BMP2 promoter within the first 6mA peak region to regulate its transcription. Under Pi stimulation, BMP2 activity was upregulated with the Oct4 (pGL3-Oct4-WT) but not with the mutation. In addition, this increase could be dramatically antagonized by ALKBH1 knockdown (Figure 8I). Taken together, 6mA demethylation by ALKBH1 promotes the binding of Oct4 to the BMP2 promoter, thus increasing the transcriptional activity of BMP2.
Oct4 mediates the regulation of ALKBH1 on BMP2.
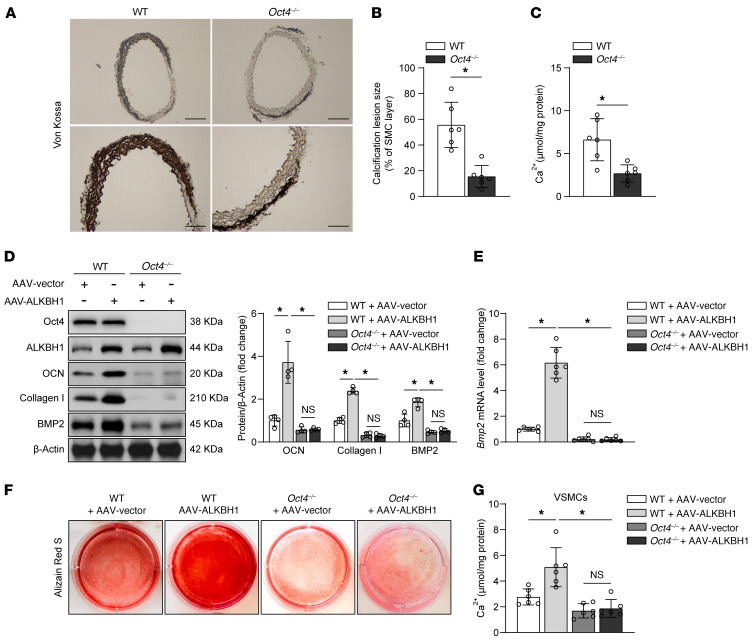
To explore whether Oct4 is involved in the regulation of ALKBH1 on BMP2, we used aorta and primary cells from tamoxifen-induced Oct4 knockout mice. Aortic rings from Oct4WT/WT-Myh11-Cre/ERT2 (WT) and Oct4F/F-Myh11-Cre/ERT2 (Oct4–/–) mice were cultured in an osteogenic medium for 14 days. Von Kossa staining (Figure 9, A and B) and calcium content quantification (Figure 9C) results showed decreased calcification in Oct4–/– mice. This observation was reinforced by transfection AAV-Vector or AAV-ALKBH1 into calcified primary VSMCs from WT or Oct4–/– mice. Depletion of Oct4 significantly alleviated the procalcifying effect of ALKBH1 overexpression by downregulating BMP2 protein and mRNA expressions (Figure 9, D–G). Based on these findings, we concluded that Oct4 mediates the regulation of ALKBH1 on BMP2.
Figure 9. Oct4 mediates the regulation of ALKBH1 on BMP2.
(A–C) Representative von Kossa staining images (A) and quantification (B) of aortic rings from Oct4WT/WT-Myh11-Cre/ERT2 (WT) and Oct4F/F-Myh11-Cre/ERT2 (Oct4–/–) mice cultured in osteogenic medium for 14 days. Bar graphs representative of calcium content (C) from these 2 groups (n = 6 per group). Scale bar: 100 μm. (D–G) Western blot analysis (D) of BMP2, Oct4, ALKBH1, and osteogenic phenotype marker (OCN and Collagen I) expression in calcified primary VSMCs from WT or Oct4–/– mice transfected with AAV-Vector or AAV-ALKBH1. Quantitative real-time PCR analysis of Bmp2 expression in all of the experimental cohorts (E). Alizarin red staining (F) and calcium content quantification (G) performed in all of the groups for detecting calcification formation (n = 4–6 per group). Statistical significance was assessed using 2-tailed t tests (B and C) and 1-way ANOVA followed by Bonferroni’s test (D–G). All values are presented as mean ± SD. *P < 0.05.
Discussion
The heterogeneity of VSMCs and their tight communication with the bloodstream render it challenging to uncover the mechanism underlying VC (1). In the current study, we found that the DNA 6mA levels in leukocytes and VSMCs were dynamically decreased along with calcification progression in CKD. This suggests a potential role of DNA 6mA modification in calcification formation. Upregulation of ALKBH1, but not N6AMT1, mediates this observed 6mA reduction. Moreover, in vivo gain- and loss-of-function experiments showed that ALKBH1 deficiency in VSMCs bestows a strong anticalcification effect, while exogenous overexpression of ALKBH1 augmented calcification formation. Mechanistically, ALKBH1-demethylated DNA 6mA could facilitate Oct4 binding to BMP2 promoter and subsequently increase BMP2 transcription, which resulted in osteogenic reprogramming of VSMCs and VC progression. Taken together, these results demonstrate that the ALKBH1 acts as a catastrophic trigger for VC.
Mounting evidence supports a critical role of DNA methylation and demethylation in VSMCs phenotypic transition due to environmental signal modulation (32). Previous research has focused mainly on canonical 5mC due to its abundance and significance in eukaryotes. A set of VSMC contractile and osteogenic genes, including klotho (12), TAGLN (15), and peroxisome proliferator-activated receptor gamma (33) are regulated by DNA 5 mC methylation, which could be consequently associated with VSMC phenotypic transition and pathogenesis of VC. In addition to the genome-wide DNA 5mC, ALKBH1-mediated 6mA DNA demethylation was recently identified in human cardiovascular diseases (19). However, it remains unknown whether VSMCs can adopt novel epigenetic mechanisms that are rarely used in normal human tissues during VC. Here, we took advantage of the ELISA to quantify the level of DNA 6mA modification in vivo and in vitro. Previous epigenome-wide association studies (EWASs) based on whole-blood DNA methylation have identified numerous CpG sites that are associated with aging (34) and all-cause mortality (35). In this study, we demonstrated that 6mA levels were markedly decreased in leukocytes and radial arteries from patients with CKD with VC, as well as the CKD mice.
Human and murine leukocyte DNA 6mA levels measured in our study were approximately 0.04% of the total adenines, which is comparable with previous research (17). Our findings suggested that epigenetic DNA 6mA modifications occurred systemically in response to the CKD-induced hyperphosphatemia, both in circulating leukocytes and in situ VSMCs. Moreover, 6mA levels in the CKD mouse model exhibited a similar decreasing pattern with humans. Therefore, leukocyte DNA 6mA modification levels, at least in part, may reflect the corresponding systematic change of 6mA modification in VSMCs. Calcium deposition in the arterial wall is considered a typical feature for VC. As we observed, the calcification Agatston score and Volume score of aortic arch exhibited a negative and positive correlation with 6mA and ALKBH1 levels in leukocytes from patients with CKD, respectively. As the severity of calcification increases, 6mA levels narrow between different stages of VC progression. These interpretations are limited, as the findings in CKD patients with VC are observational due to insufficient access to direct tissue artery biopsies. Despite these limitations, our preclinical studies provide at least preliminary indications that DNA 6mA levels may function as a novel epigenetic biomarker for early diagnosis of VC initiation, since regular MDCT does not have the sensitivity to identify minor aortic calcification lesion. Leukocyte isolation and analysis could be a simple yet highly effective clinical approach since MDCT contrast agents may have adverse effects on uremic patients.
Gain- and loss-of-function experiments corroborated the evidence for causal ALKBH1 involvement in VC. ALKBH1 deficiency in VSMCs significantly alleviated the accumulation of calcium deposition in the arterial wall, whereas ALKBH1 overexpression exacerbated this pathological process. Specific mediators such as SOX9, DLX5, RUNX2, and BMP2 are involved in VSMC osteogenic reprogramming and calcification development (36). Here, we found that BMP2 expression was significantly blunted by ALKBH1 deficiency in calcified VSMCs. Moreover, the notable procalcifying outcome arising from ALKBH1 overexpression was abolished upon BMP2 depletion. That said, ALKBH1-mediated VC is dependent on BMP2. However, the mechanism by which ALKBH1 downregulates VSMC contractile markers under high phosphate stimulation is not fully understood. Interestingly, previous work found that BMP2 could repress TAGLN promoter activity in VSMCs via a common cis-acting transforming growth factor control element, which is present in promoter regions of multiple contractile VSMC genes (37).
Thus, a critical question to consider is how ALKBH1 regulates BMP2 expression. In mouse embryonic stem cells (mESCs), 6mA was enriched at young long interspersed nuclear element 1 transposon, and its deposition correlates with epigenetic silencing (38). Recent studies have also shown that 6mA affects gene expression by regulating the binding of a specific transcription factor to the target gene promoter region (18, 39). Our results revealed that Pi induced ALKBH1-demethylated 6mA on an Oct4 binding region within the BMP2 promoter, which facilitated Oct4 binding and promoted BMP2 transcriptional activation. Although the Oct4 binding motif has multiple adenosines that are required for Oct4 binding, we acknowledge that a limitation of the current study is that we could not precisely identify which adenosine undergoes N6 methylation. Recent sequencing technology could not distinguish genomic 6mA and adenosine. In addition, application of VSMC-specific Alkbh1 gene knockout mice will be better to bolster our findings.
In summary, our clinical investigation established an association of dynamically decreased DNA 6mA modification with the progression of VC. Our mechanistic results demonstrated the procalcification impact of ALKBH1-demethylated DNA 6mA modification in CKD. From a clinical standpoint, these observations warrant further study of DNA 6mA level as an early diagnosis approach and ALKBH1 inhibition as a potential therapeutic strategy for the prevention of VC.
Methods
Study population and patient artery samples.
This case control study was conducted from March 2018 to January 2020 in the Donghua Hospital of Sun Yat-sen University, China. A total of 198 patients diagnosed with CKD were recruited in our study. We excluded 10 patients who met the exclusion criteria and 15 patients who missed the data of MDCT or plasma biomedical tests. Clinical and biochemical parameters were collected from the electronic medical record in the hospital.
The diagnosis of CKD was based on eGFR decline present for 6 months or longer. CKD stage 1 to 2 was defined as albumin/creatinine ratio (ACR) (mg/g) ≥ 17 in men and ≥ 25 in women, and an eGFR ≥ 60 mL/min per 1.73 m2 following the Kidney Disease Outcomes Quality Initiative (K/DOQI) guidelines (40). CKD stage 3 to 5 was defined as an estimated GFR < 60 mL/min per 1.73 m2 among study participants before receiving dialysis. Patients were excluded for the following: (a) age < 18 and ≥ 90 years; (b) pregnant; (c) administration of any medication as phosphate binders, vitamin D analogs, or the corticosteroids within 3 months before recruitment; (d) acute renal failure; (e) uncontrolled hyperlipidemia, and (f) history of hepatic diseases or neoplastic disease.
A 4 to 6 mm segment of the radial artery was removed from patients with CKD who underwent an arterial venous fistula operation with diagnosed aortic arch calcification (CKD, n = 10) or from patients who underwent amputation surgery due to upper limb trauma without diagnosed CKD or diabetes mellitus (control, n = 8). Fat and adventitia were removed from samples before storing them in liquid nitrogen.
Assessment of calcification score.
Among the cohort, all patients underwent a chest MDCT scan with standard electrocardiographically gated (ECG-gated) protocol to evaluate aortic arch calcification. All the Agatston and Volume scores of images were blindly quantified by 2 independent investigators with Siemens Syngo CT Workplace software according to standard criteria (41). The aortic arch refers to the part between the ascending and descending aorta. To measure the calcification scores, the CT images were reconstructed with 1 mm–thick slices. The presence of calcification was defined as an Agatston score or Volume score greater than 0 in the present study. Studies focused on coronary artery describing Agatston scores greater than 400 as severe calcification (42–44). Considering the higher calcification scores in aortic arch (Agatston score [median 411]; Volume score [median 461]) than coronary artery (Agatston score [median 163]; Volume score [median 283]), patients were divided into non-VC (Agatston score or Volume score = 0), mild (500 ≥ Agatston score or Volume score > 0), or severe (Agatston score or Volume score > 500) subgroups as previously described (45). Calcification scores were adjusted with a calcium standard calibration phantom that was scanned along with the participant (46). In addition, calcification scores and the relevant statistical analyses were adjusted for age, sex, and ethnicity (47).
Laboratory analyses.
An overnight fasting median cubital venous blood was collected for analyses of biochemical parameters from patients with CKD before dialysis. Leukocyte cells were isolated for subsequent DNA and RNA analyses. Plasma levels of potassium, calcium, phosphate, blood urea nitrogen (BUN), creatinine (CREA), uric acid (URCA), albumin (ALB), alanine transaminase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), cholesterol (CHOL), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) were analyzed using the standard certified TBA-120 auto-analyzer (Toshiba Medical Systems) in the Central Clinical Laboratory at Donghua Hospital of Sun Yat-sen University. The suPAR in plasma was measured by the detection kit (ViroGates) according to the manufacturer’s instructions. The estimated glomerular filtration rate was assessed using the modified Modification of Diet in Renal Disease (MDRD) equation for the Chinese population (48).
Quantification of 6mA DNA methylation.
The DNeasy Blood & Tissue Kit from Qiagen was used to extract genomic DNA from peripheral blood specimens of humans and mice and cultured cell lines. The integration of genomic DNA was confirmed on an agarose gel, and the concentration was measured by a NanoDrop spectrophotometer. The 6mA DNA methylation level was quantified using Methyl Flash 6mA DNA Methylation ELISA Kit (Colorimetric) from Epigentek following the manufacturer’s protocol. In brief, the methylated fraction of a total of 100 ng genomic DNA was recognized by a 6mA antibody and quantified by an ELISA-like reaction. The percentage of 6mA (6mA %) was calculated by optical density (OD) value according to the recommended formula. Methylated and unmethylated DNA were incubated in each test as positive and negative controls, respectively. Each sample ran in duplicate.
Chemicals and antibodies.
Adenine (A8626), β-GP (G9422), and l-ascorbic acid (A4544) were purchased from Sigma-Aldrich. For Western blot (WB), immunofluorescence (IF), and immunohistochemistry (IHC), antibodies against RUNX2 (12556S; 1:1000 for WB), SOX9 (82630S; 1:1000 for WB), and Calponin1 (17819S; 1:1000 for WB) were obtained from Cell Signaling Technology. Antibodies against α-SMA (ab7817; 1:2000 for WB; 1:100 for IF), BMP2 (ab14933; 1:1000 for WB; 1:100 for IF), SM22α (ab14106; 1:2000 for WB), OPN (ab8448; 1:1000 for WB), and Oct4 (ab19857; 1:500 for WB) were purchased from Abcam. Antibodies against Collagen I (NB600-408; 1:1000 for WB) were purchased from Novus. Antibodies against ALKBH1 (OACA06689; 1:1000 for WB; 1:100 for IF and IHC) and N6AMT1 (OAAN02175; 1:1000 for WB; 1:100 for IF and IHC) were obtained from Aviva Systems Biology. Antibodies against 6mA (ABE572; 1:100 for IF and IHC) for ChIP were obtained from EMD Millipore, and Oct4 (2750S) for ChIP was purchased from Cell Signaling Technology. The OCN (sc-390877; 1:500 for WB), DXL5 (sc-398150; 1:500 for WB), GAPDH (sc-32233; 1:1000 for WB), and β-Actin (sc-47778; 1:1000 for WB) antibodies were purchased from Santa Cruz Biotechnology.
Animal experiments.
Eight-week-old male C57BL/6J mice were purchased from Laboratory Animal Center of Sun Yat-sen University. Oct4F/F (Pou5f1tm1Scho) and Myh11-Cre/ERT2 mice were purchased from the Jackson Laboratory to generate Oct4F/F-Myh11-Cre/ERT2 mice. Cre-mediated recombination was achieved via 10 daily intraperitoneal injections of tamoxifen (MilliporeSigma) at 6 weeks of age. Male Oct4WT/WT-Myh11-Cre/ERT2 mice were referred to as WT, and tamoxifen-injected male Oct4F/F-Myh11-Cre/ERT2 mice were referred to as Oct4–/– for simplicity. All mice were housed in a temperature-controlled environment under a 12-hour light cycle, and they had free access to water and the assigned diet. For the adenine diet–induced CKD model, mice were randomly provided with a chow diet as the control group, or a special diet containing 0.2% adenine, 1.2% phosphorus as the CKD group (49–52). For the 5/6 nephrectomy–induced CKD model, we followed a modified protocol based on the well-established 2-step procedure we used in rats (53–55). Briefly, anesthesia was induced in 8-week-old male mice with isoflurane (induction 5%, maintenance 2%). First, the upper and lower poles of the left kidney were ligated and excised under a microscope. Two-thirds of the left kidney was removed. One week later, mice were subjected to removal of right kidney to achieve 5/6 nephrectomy. Four weeks after initiation of the special diet or first-step surgery, mice were injected with the indicated virus (5 × 109 plaque-forming units per kilogram per mouse) via tail vein, respectively, as previously described (56, 57). Four weeks later, mice were fasted overnight and weighted before euthanasia, and blood was collected and separated into plasma and leukocytes. Blood levels of fast blood glucose, phosphorus, creatinine, and urea BUN were measured by autoanalyzer (Hitachi). Plasma cholesterol and triglyceride were measured using specific reagents (Infinity, Thermo Fisher Scientific). Plasma levels of calcium were measured using the detection kit (Abcam, ab102505). Plasma levels of ALP were measured using the QuantiChrom Alkaline Phosphatase Assay Kit (catalog DALP-250) from Bioassay Systems. The suPAR in plasma was measured by detection kit (R&D Systems, DY531) according to the manufacturer’s instructions. Whole aortas were harvested and carefully dissected for further analyses.
Blood pressure.
Measurements of systolic blood pressure (SBP) and diastolic blood pressure (DBP) in mice were performed by noninvasive tail cuff plethysmography (CODA, Kent Scientific Corporation), before (W0), 2 (W2), and 4 (W4) weeks after virus injection. These measurements were conducted in conscious and trained mice in the morning with 5 acclimation cycles followed by 15 measurement cycles. The mean value of measurements was used for comparison.
Immunohistochemistry.
Vascular sections (5 μm) were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked using 0.3% H2O2 at room temperature for 20 minutes. Heat-mediated antigen retrieval was performed using 10% citrate buffer. Sections were stained with primary antibodies overnight at 4°C followed with EnVision+ Dual Link System-HRP (Dako) and visualized with diaminobenzidine. OLYMPUS BX51 microscope (Japan) was used for image collecting, and the scores were calculated by the percentages of positive cells and staining intensities as previously described (58).
Immunofluorescence.
Slides (8 μm) were washed with diluted water and fixed in iced acetone for 10 minutes and then blocked with goat serum. Slides were incubated with antibodies at 4°C overnight and then incubated with Alexa Fluor (Jackson ImmunoResearch) secondary antibodies for 1 hour at 37°C. Nuclei were stained with DAPI. Images were acquired by Olympus fluorescence microscope and quantified by Image-Pro Plus 6.0 (Media Cybernetics).
Histological examination.
Kidney was fixed in 10% formalin and embedded in paraffin. Sagittal sections were stained with hematoxylin and eosin (H&E) for morphology using a solution from Sigma-Aldrich, Sirus red for fibrosis using stain kit (Polysciences, 24901), and von Kossa staining for mineral deposition (American MasterTech Scientific, KTVKO) according to the manufacturers’ instructions.
Ex vivo cell and vascular tissue culture.
Primary murine VSMCs were isolated from aortas of 8-week-old male C57BL/6J mice, 12-week-old male Oct4WT/WT-Myh11-Cre/ERT2 mice, or tamoxifen-pretreated 12-week-old male Oct4F/F-Myh11-Cre/ERT2 mice, and cultured in a growth medium as described by Hou et al (59). Human aortic smooth muscle cells (HASMCs) were purchased from the American Type Culture Collection and cultured in M231 medium (Life Technology) containing 10% fetal bovine serum (FBS) (Gibco). All experiments were performed with VSMCs at passages 3 to 6. Mice aortic arteries were dissected under sterile conditions from 8-week-old male C57BL/6J mice, 12-week-old male Oct4WT/WT-Myh11-Cre/ERT2 mice, or tamoxifen-pretreated 12-week-old male Oct4F/F-Myh11-Cre/ERT2 mice. After removing the adventitia, the vessels were cut into 2 to 3 mm rings and placed in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) containing 10% FBS.
Cells were transfected with control siRNA, ALKBH1, or OCT4 siRNA (Santa Cruz Biotechnology), accordingly, and with Lipofectamine RNAiMAX Reagent (Invitrogen) as we described (60). Plasmids were transfected into HASMCs by electroporation using the P1 Primary Cell 4D-Nucleofector X Kit L from Lonza following the manufacturer’s instruction. For indicated viruses, cells were seeded overnight prior to viral infections. The efficiency of transfection was determined by real-time polymerase chain reaction (PCR) or Western blot.
In vivo calcification quantification.
Calcification was identified by von Kossa staining and analyzed by ImageJ (NIH). Aortic sections were treated with 5% silver nitrate and exposed to ultraviolet light for 30 minutes, rinsed, and incubated with 5% sodium thiosulfate. Calcified spots were stained brown. The aortic arch was decalcified with 0.6 mmol/L hydrochloric acid at 37°C for 48 hours. Released calcium was quantified using the Calcium Assay Kit (Abcam, ab102505) in compliance with the manufacturer’s protocol. The content of calcium was normalized by the dry weight of vascular tissues. The presence of calcification was defined by positive von Kossa staining in the medial aortic layer as calcification lesion size (% of SMC layer) greater than 0 in this study. Mice were divided into non-VC (calcification lesion size = 0), mild (10% ≥ calcification lesion size > 0), or severe (calcification lesion size > 10%) subgroups.
In vitro calcification quantification.
Cells or aortic rings were cultured in an osteogenic medium containing 0.25 mmol/L l-ascorbic acid and 10 mmol/L β-glycerophosphate for 14 days to induce in vitro calcification. Calcification was determined by Alizarin red stain (Thermo Fisher Scientific, AA42746AP) as previously described (60). Positively stained cells showed a red color. The calcium deposition in the plate was decalcified and subjected to colorimetric analysis using a Calcium Assay Kit (Abcam, ab102505) and calibrated to the protein content (60). In parallel duplicate, ALP activity was measured using an ALP assay kit (QuantiChrom, DALP-250) and normalized to the levels of total protein according to the manufacturer’s instructions.
Quantitative real-time PCR.
Total RNA from peripheral leukocytes, cultured cells, or murine aortas were extracted with TRIzol (Invitrogen) and reverse-transcribed into cDNA with an iScript cDNA Synthesis Kit (Bio-Rad). PCR amplification was performed using Bio-Rad SYBR Green on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The relative mRNA levels were calibrated by the housekeeping gene GAPDH. The primers are listed in Supplemental Table 4.
Western blotting analysis.
Cell lysates or aortic homogenates were performed by Western blot analysis as previously described (60). The intensity of the individual band was quantified by densitometry using ImageJ (NIH).
Chromatin immunoprecipitation (ChIP)–qPCR.
The bioinformatics screening of functional 6mA binding sites was obtained from the NCBI GEO Data Sets (GSE118093). ChIP analyses were performed using the Simple Chip Enzymatic Chromatin immunoprecipitation kit (Cell Signaling Technology, 9002S) following the manufacturer’s protocols. Briefly, treated cells were crosslinked with formaldehyde and then quenched by glycine, and finally digested with micrococcal nuclease. The lysates were immunoprecipitated with 5 μg rabbit anti-IgG, anti-6mA, or anti-Oct4 antibody overnight at 4°C, followed by incubation with 50 μL ChIP-Grade Protein G Agarose beads (Cell Signaling Technology, 9007S) for 2 hours at 4°C with gentle rotation. The crosslinked DNA fragments were eluted, reversed, and purified. ChIP DNA was amplified by real-time PCR with designed primers (Supplemental Table 4). A 2% amount of each reaction was used as an input reference. The results were analyzed by the following formula: percentage of input = 2% × 2(Ct 2% input sample – Ct IP sample).
Recombinant adeno-associated virus construction.
To generate AAV encoding mouse Alkbh1 (NM_001102565.1), the genomic AAV vector, AAV helper plasmid, and AAV serotype 2 plasmid (Origene) were cotransfected into HEK293 cells (American Type Culture Collection), and the harvested viral particles were purified as previously outlined (59, 61). Double digestion was done with SgfI and MluI restriction enzymes (Sigma-Aldrich) to remove the Alkbh1 open reading frame clone from pCMV6-Myc-DDK-Alkbh1 plasmid (Origene, MR222060) to construct genomic AAV vector. In order to construct optimal AAV expressing shRNA against Alkbh1 and Bmp2, 3 lines of the corresponding AAV were modified and packaged and then transfected into murine VSMCs to determine the efficiency of knockdown (Supplemental Figure 11). The optimal sequences were listed as follows: Alkbh1 shRNA with the sense oligonucleotides 5′-CCGGGAAATACTCAGCAGATCATTACTCGAGTAATGATCTGCTGAGTATTTCTTTTTG-3′; and Bmp2 shRNA with the sense oligonucleotides 5′-CCGGCCTCCGGGCTATCATGCCTTTCTCGAGAAAGGCATGATAGCCCGGAGGTTTTT-3′. To ensure the restricted genomic manipulation in the smooth muscle cell, all the AAV vectors were ligated with Tagln promoter.
Plasmid construction and luciferase reporter assay.
The serial DNA fragments from the human BMP2 promoter (–3319 bp) were amplified by PCR. The PCR product was digested with SacI and HindIII (Sigma-Aldrich) and cloned into pGL3-Vector to generate the BMP2 luciferase reporter constructs. Mutant constructs of the deleted binding site for Oct4 in the BMP2 promoter were generated by designed primers. The constructs were verified by sequencing. After the indicated treatment, the reporter plasmid was cotransfected with internal control Renilla luciferase plasmid (Promega) into HASMCs by electroporation. Transcriptional activity was detected by the Dual-Luciferase Reporter Assay Kit obtained from Promega according to the manufacturer’s instructions. The related primers are listed in Supplemental Table 4.
Lentivirus transfection.
HASMCs were infected with recombinant lentivirus expressing control shRNA (Santa Cruz Biotechnology, sc-108080), ALKBH1 shRNA (Santa Cruz Biotechnology, sc-60153-V) according to the manufacturer’s instruction. Colonies were selected by treatment with 5 μg/mL puromycin (Sigma-Aldrich) for 7 days.
Statistics.
All analyses were performed with SPSS 23.0 software (SPSS Inc.). Values are expressed as mean ± standard deviation. For comparisons between 2 groups, significance was determined using Student’s t test or nonparametric Mann-Whitney U test. For comparisons among multiple groups, ANOVA was performed, followed by the post hoc Bonferroni’s or Dunnett’s test. The statistical significance of correlations was assessed by Pearson’s correlation coefficient analysis. Two-tailed P < 0.05 (indicated by *, #, or †) was considered statistically significant.
Study approval.
The protocol for clinical study conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the Donghua Hospital of Sun Yat-sen University. All participants signed informed consent before entering this study. Written informed consent was provided for pictures appearing in the manuscript. The experimental animal protocols were reviewed and approved by the Ethics Committee of Zhongshan School of Medicine, Sun Yat-sen University.
Author contributions
LO and HH designed the research. LO, MZ, and WL performed the experiments. LO, XS, LT, YZ, PZ, CX, and JC analyzed and interpreted the data. LO and HH wrote the manuscript with comments from all authors. All authors approved the final version of the manuscript. The order of co–first authors was determined by their efforts and contributions to the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (8201101103, 81870506, and 81670676), Project of Traditional Chinese Medicine in Guangdong Province (20201062), Basic Research Project of Shenzhen Science and Technology Innovation Committee (JCYJ20180306174648342 and JCYJ20190808102005602), Shenzhen Futian District Public Health Research Project (FTWS2019003), and Shenzhen Key Medical Discipline Construction Fund (SZXK002) to HH. In addition, this work was supported by the National Natural Science Foundation of China (82073408 to JC) and Dongguan Social Science and Technology Development Project (2018507150461629 to XS).
Version 1. 05/18/2021
In-Press Preview
Version 2. 07/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(14):e146985.https://doi.org/10.1172/JCI146985.
See the related Commentary at ALKBH1 reduces DNA N6-methyladenine to allow for vascular calcification in chronic kidney disease.
Contributor Information
Liu Ouyang, Email: ouyliu@mail2.sysu.edu.cn.
Xiaoyan Su, Email: suxiaoyan769@hotmail.com.
Wenxin Li, Email: liwenx5@mail2.sysu.edu.cn.
Liangqiu Tang, Email: liangqiut@126.com.
Mengbi Zhang, Email: zmb951025@163.com.
Yongjun Zhu, Email: zhuyj53@mail2.sysu.edu.cn.
Changming Xie, Email: xiechm3@gmail.com.
Puhua Zhang, Email: zhangph6@126.com.
Jie Chen, Email: 1450517759@qq.com.
Hui Huang, Email: huangh8@mail.sysu.edu.cn.
References
- 1.Blaha MJ, et al. Associations between C-reactive protein, coronary artery calcium, and cardiovascular events: implications for the JUPITER population from MESA, a population-based cohort study. Lancet. 2011;378(9792):684–692. doi: 10.1016/S0140-6736(11)60784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Timmis A, et al. European Society of Cardiology: cardiovascular disease statistics 2017. Eur Heart J. 2018;39(7):508–579. doi: 10.1093/eurheartj/ehx628. [DOI] [PubMed] [Google Scholar]
- 3.Moe SM, Chen NX. Mechanisms of vascular calcification in chronic kidney disease. J Am Soc Nephrol. 2008;19(2):213–216. doi: 10.1681/ASN.2007080854. [DOI] [PubMed] [Google Scholar]
- 4.Guerin AP, et al. Impact of aortic stiffness attenuation on survival of patients in end-stage renal failure. Circulation. 2001;103(7):987–992. doi: 10.1161/01.CIR.103.7.987. [DOI] [PubMed] [Google Scholar]
- 5.Pursnani A, et al. Guideline-based statin eligibility, coronary artery calcification, and cardiovascular events. JAMA. 2015;314(2):134–141. doi: 10.1001/jama.2015.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanzer P, et al. Medial vascular calcification revisited: review and perspectives. Eur Heart J. 2014;35(23):1515–1525. doi: 10.1093/eurheartj/ehu163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iribarren C, et al. Calcification of the aortic arch: risk factors and association with coronary heart disease, stroke, and peripheral vascular disease. JAMA. 2000;283(21):2810–2815. doi: 10.1001/jama.283.21.2810. [DOI] [PubMed] [Google Scholar]
- 8.Opdebeeck B, et al. Indoxyl sulfate and p-Cresyl sulfate promote vascular calcification and associate with glucose intolerance. J Am Soc Nephrol. 2019;30(5):751–766. doi: 10.1681/ASN.2018060609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shroff R, et al. Mechanistic insights into vascular calcification in CKD. J Am Soc Nephrol. 2013;24(2):179–189. doi: 10.1681/ASN.2011121191. [DOI] [PubMed] [Google Scholar]
- 10.Salazar VS, et al. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12(4):203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. BMP-2 promotes phosphate uptake, phenotypic modulation, and calcification of human vascular smooth muscle cells. Atherosclerosis. 2008;199(2):271–277. doi: 10.1016/j.atherosclerosis.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, et al. Indoxyl sulfate enhance the hypermethylation of klotho and promote the process of vascular calcification in chronic kidney disease. Int J Biol Sci. 2016;12(10):1236–1246. doi: 10.7150/ijbs.15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pons D, et al. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30(3):266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 14.Goettsch C, et al. MicroRNA in cardiovascular calcification: focus on targets and extracellular vesicle delivery mechanisms. Circ Res. 2013;112(7):1073–1084. doi: 10.1161/CIRCRESAHA.113.300937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montes de Oca A, et al. High-phosphate-induced calcification is related to SM22alpha promoter methylation in vascular smooth muscle cells. J Bone Miner Res. 2010;25(9):1996–2005. doi: 10.1002/jbmr.93. [DOI] [PubMed] [Google Scholar]
- 16.Zhu S, et al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 2018;28(7):1067–1078. doi: 10.1101/gr.231068.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao CL, et al. N(6)-Methyladenine DNA modification in the human genome. Mol Cell. 2018;71(2):306–318. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, et al. Association of N(6)-methyladenine DNA with plaque progression in atherosclerosis via myocardial infarction-associated transcripts. Cell Death Dis. 2019;10(12):909. doi: 10.1038/s41419-019-2152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, et al. DNA N(6)-methyladenine modification in hypertension. Aging (Albany NY) 2020;12(7):6276–6291. doi: 10.18632/aging.103023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou C, et al. DNA N(6)-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res. 2016;4:16033. doi: 10.1038/boneres.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie SA, et al. Matrix stiffness determines the phenotype of vascular smooth muscle cell in vitro and in vivo: role of DNA methyltransferase 1. Biomaterials. 2018;155:203–216. doi: 10.1016/j.biomaterials.2017.11.033. [DOI] [PubMed] [Google Scholar]
- 22.Hadji F, et al. Altered DNA METHYLATION OF LONG NONcoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation. 2016;134(23):1848–1862. doi: 10.1161/CIRCULATIONAHA.116.023116. [DOI] [PubMed] [Google Scholar]
- 23.Azam TU, et al. Soluble urokinase receptor (SuPAR) in COVID-19-related AKI. J Am Soc Nephrol. 2020;31(11):2725–2735. doi: 10.1681/ASN.2020060829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahm E, et al. Bone marrow-derived immature myeloid cells are a main source of circulating suPAR contributing to proteinuric kidney disease. Nat Med. 2017;23(1):100–106. doi: 10.1038/nm.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayek SS, et al. A tripartite complex of suPAR, APOL1 risk variants and αvβ3 integrin on podocytes mediates chronic kidney disease. Nat Med. 2017;23(8):945–953. doi: 10.1038/nm.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayek SS, et al. Soluble urokinase receptor and acute kidney injury. N Engl J Med. 2020;382(5):416–426. doi: 10.1056/NEJMoa1911481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayek SS, et al. Soluble urokinase receptor and chronic kidney disease. N Engl J Med. 2015;373(20):1916–1925. doi: 10.1056/NEJMoa1506362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ougland R, et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. 2012;30(12):2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rong S, et al. Vascular calcification in chronic kidney disease is induced by bone morphogenetic protein-2 via a mechanism involving the Wnt/β-catenin pathway. Cell Physiol Biochem. 2014;34(6):2049–2060. doi: 10.1159/000366400. [DOI] [PubMed] [Google Scholar]
- 30.Xie Q, et al. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018;175(5):1228–1243. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poetsch AR, Plass C. Transcriptional regulation by DNA methylation. Cancer Treat Rev. 2011;37(Suppl 1):S8–12. doi: 10.1016/j.ctrv.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu Rev Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, et al. High phosphate-induced downregulation of PPARγ contributes to CKD-associated vascular calcification. J Mol Cell Cardiol. 2018;114:264–275. doi: 10.1016/j.yjmcc.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. doi: 10.1038/ncomms14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson B, Towler DA. Arterial calcification and bone physiology: role of the bone-vascular axis. Nat Rev Endocrinol. 2012;8(9):529–543. doi: 10.1038/nrendo.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King KE, et al. Krüppel-like factor 4 (KLF4/GKLF) is a target of bone morphogenetic proteins and transforming growth factor beta 1 in the regulation of vascular smooth muscle cell phenotype. J Biol Chem. 2003;278(13):11661–11669. doi: 10.1074/jbc.M211337200. [DOI] [PubMed] [Google Scholar]
- 38.Wu TP, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532(7599):329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, et al. Epigenetic methylations on N6-adenine and N6-adenosine with the same input but different output. Int J Mol Sci. 2019;20(12):E2931. doi: 10.3390/ijms20122931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.National Kidney Foundation K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(2 Suppl 1):S1–266. [PubMed] [Google Scholar]
- 41.Agatston AS, et al. Quantification of coronary artery calcium using ultrafast computed tomography. J Am Coll Cardiol. 1990;15(4):827–832. doi: 10.1016/0735-1097(90)90282-T. [DOI] [PubMed] [Google Scholar]
- 42.Budoff MJ, et al. Testosterone treatment and coronary artery plaque volume in older men with low testosterone. JAMA. 2017;317(7):708–716. doi: 10.1001/jama.2016.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budoff MJ, et al. Prognostic value of coronary artery calcium in the PROMISE study (prospective multicenter imaging study for evaluation of chest pain) Circulation. 2017;136(21):1993–2005. doi: 10.1161/CIRCULATIONAHA.117.030578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raggi P, et al. Identification of patients at increased risk of first unheralded acute myocardial infarction by electron-beam computed tomography. Circulation. 2000;101(8):850–855. doi: 10.1161/01.CIR.101.8.850. [DOI] [PubMed] [Google Scholar]
- 45.Odink AE, et al. Association between calcification in the coronary arteries, aortic arch and carotid arteries: the Rotterdam study. Atherosclerosis. 2007;193(2):408–413. doi: 10.1016/j.atherosclerosis.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Nelson JC, et al. Measuring coronary calcium on CT images adjusted for attenuation differences. Radiology. 2005;235(2):403–414. doi: 10.1148/radiol.2352040515. [DOI] [PubMed] [Google Scholar]
- 47.McClelland RL, et al. Distribution of coronary artery calcium by race, gender, and age: results from the Multi-Ethnic Study of Atherosclerosis (MESA) Circulation. 2006;113(1):30–37. doi: 10.1161/CIRCULATIONAHA.105.580696. [DOI] [PubMed] [Google Scholar]
- 48.Ma YC, et al. Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol. 2006;17(10):2937–2944. doi: 10.1681/ASN.2006040368. [DOI] [PubMed] [Google Scholar]
- 49.Durlacher-Betzer K, et al. Interleukin-6 contributes to the increase in fibroblast growth factor 23 expression in acute and chronic kidney disease. Kidney Int. 2018;94(2):315–325. doi: 10.1016/j.kint.2018.02.026. [DOI] [PubMed] [Google Scholar]
- 50.Gao C, et al. Microsomal prostaglandin E synthase-1-derived PGE2 inhibits vascular smooth muscle cell calcification. Arterioscler Thromb Vasc Biol. 2016;36(1):108–121. doi: 10.1161/ATVBAHA.115.306642. [DOI] [PubMed] [Google Scholar]
- 51.Clinkenbeard EL, et al. Increased FGF23 protects against detrimental cardio-renal consequences during elevated blood phosphate in CKD. JCI Insight. 2019;4(4):123817. doi: 10.1172/jci.insight.123817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kukida M, et al. AT2 receptor stimulation inhibits phosphate-induced vascular calcification. Kidney Int. 2019;95(1):138–148. doi: 10.1016/j.kint.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, et al. Apocynin attenuates cardiac injury in type 4 cardiorenal syndrome via suppressing cardiac fibroblast growth factor-2 with oxidative stress inhibition. J Am Heart Assoc. 2015;4(7):e001598. doi: 10.1161/JAHA.114.001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng W, et al. Advanced oxidation protein products aggravate cardiac remodeling via cardiomyocyte apoptosis in chronic kidney disease. Am J Physiol Heart Circ Physiol. 2018;314(3):H475–H483. doi: 10.1152/ajpheart.00628.2016. [DOI] [PubMed] [Google Scholar]
- 55.Kong Y, et al. Statins ameliorate cholesterol-induced inflammation and improve AQP2 expression by inhibiting NLRP3 activation in the kidney. Theranostics. 2020;10(23):10415–10433. doi: 10.7150/thno.49603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lovren F, et al. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation. 2012;126(11 Suppl 1):S81–S90. doi: 10.1161/CIRCULATIONAHA.111.084186. [DOI] [PubMed] [Google Scholar]
- 57.Zhang F, et al. Long-term modifications of blood pressure in normotensive and spontaneously hypertensive rats by gene delivery of rAAV-mediated cytochrome P450 arachidonic acid hydroxylase. Cell Res. 2005;15(9):717–724. doi: 10.1038/sj.cr.7290341. [DOI] [PubMed] [Google Scholar]
- 58.Zang M, et al. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 2006;55(8):2180–2191. doi: 10.2337/db05-1188. [DOI] [PubMed] [Google Scholar]
- 59.Hou G, et al. The discoidin domain receptor tyrosine kinase DDR1 in arterial wound repair. J Clin Invest. 2001;107(6):727–735. doi: 10.1172/JCI10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang K, et al. Interleukin-18 enhances vascular calcification and osteogenic differentiation of vascular smooth muscle cells through TRPM7 activation. Arterioscler Thromb Vasc Biol. 2017;37(10):1933–1943. doi: 10.1161/ATVBAHA.117.309161. [DOI] [PubMed] [Google Scholar]
- 61.Nass SA, et al. Universal method for the purification of recombinant AAV vectors of differing serotypes. Mol Ther Methods Clin Dev. 2018;9:33–46. doi: 10.1016/j.omtm.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.