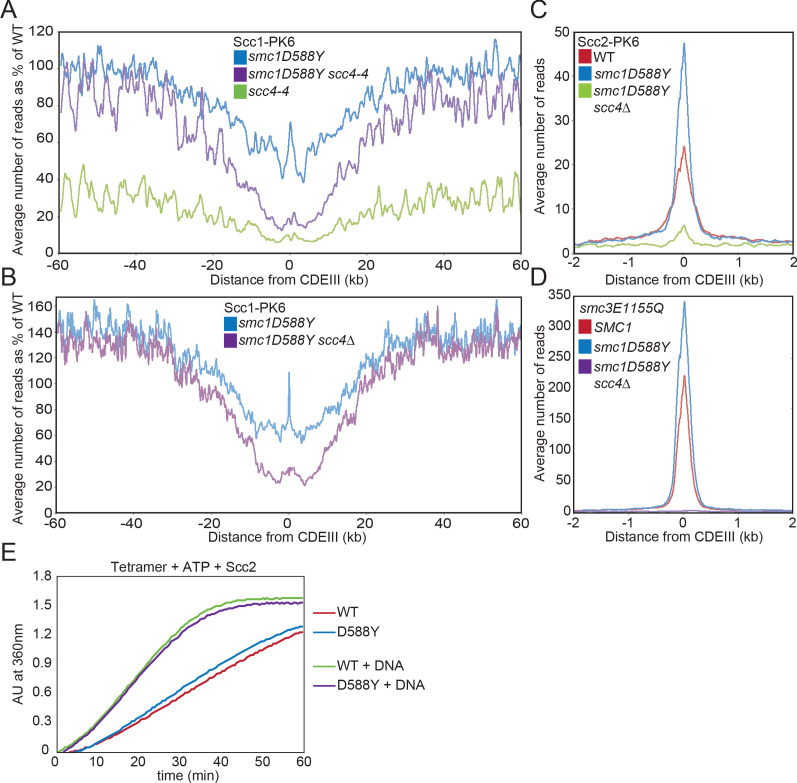
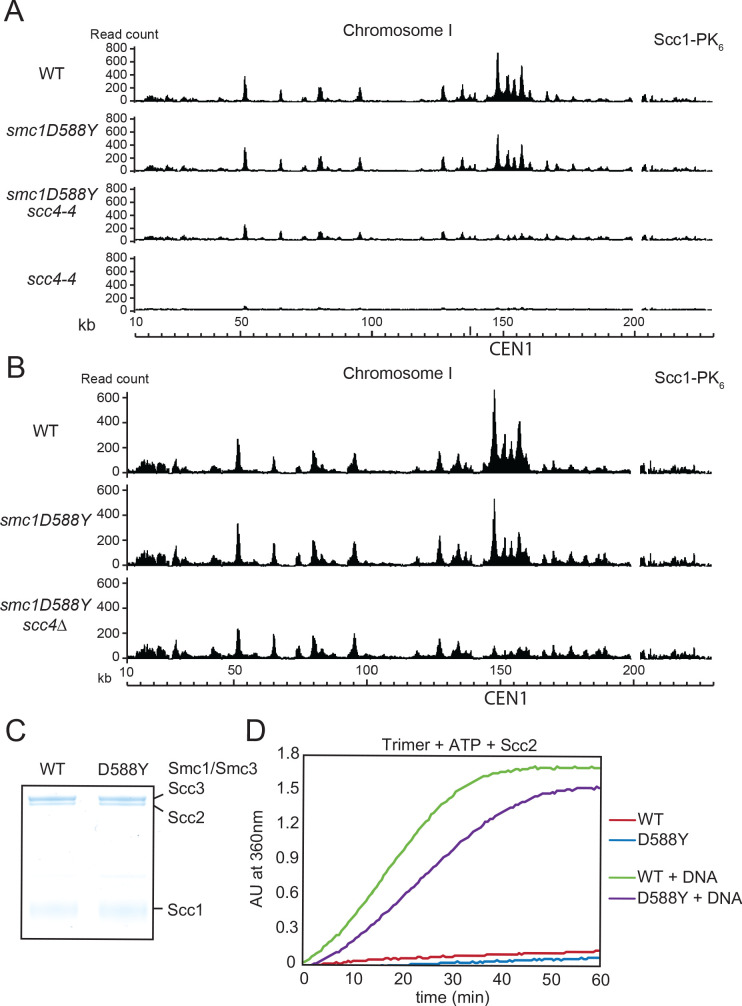
Figure 2. smc1D588Y restores cohesin occupancy on chromosome arms in the absence of Scc4.
(A) Average calibrated ChIP-seq profiles of Scc1-PK6 in smc1D588Y, scc4-4, and smc1D588Y scc4-4 cells 60 kb either side of CDEIII plotted as a percentage of the average number of reads obtained for wild-type (WT) cells. Cells were pheromone arrested in G1 at 25°C before release at 37°C into medium containing nocodazole. Samples were taken 75 min after release (K22005, K22009, K21999, K22001). (B) Average calibrated ChIP-seq profiles of Scc1-PK6 in smc1D588Y, and smc1D588Y scc4Δ cells 60 kb either side of CDEIII plotted as a percentage of the average number of reads obtained for WT cells. Cells were pheromone arrested in G1 at 25°C before release at 25°C into medium containing nocodazole. Samples were taken 60 min after release (K22005, K22009, K19624). (C) Average calibrated ChIP-seq profiles of Scc2-PK6 2 kb either side of CDEIII in cycling WT, smc1D588Y, and smc1D588Y scc4Δ cells at 25°C (K21388, K24680, K24678). (D) Average calibrated ChIP-seq profiles of ectopically expressed Smc3E1155Q-PK6 2 kb either side of CDEIII in cycling WT, smc1D588Y, and smc1D588Y scc4Δ cells at 25°C (K24562, K24689, K24564). (E) ATPase activity of WT or mutant tetramers on addition of ATP and Scc2 in the presence and absence of DNA.