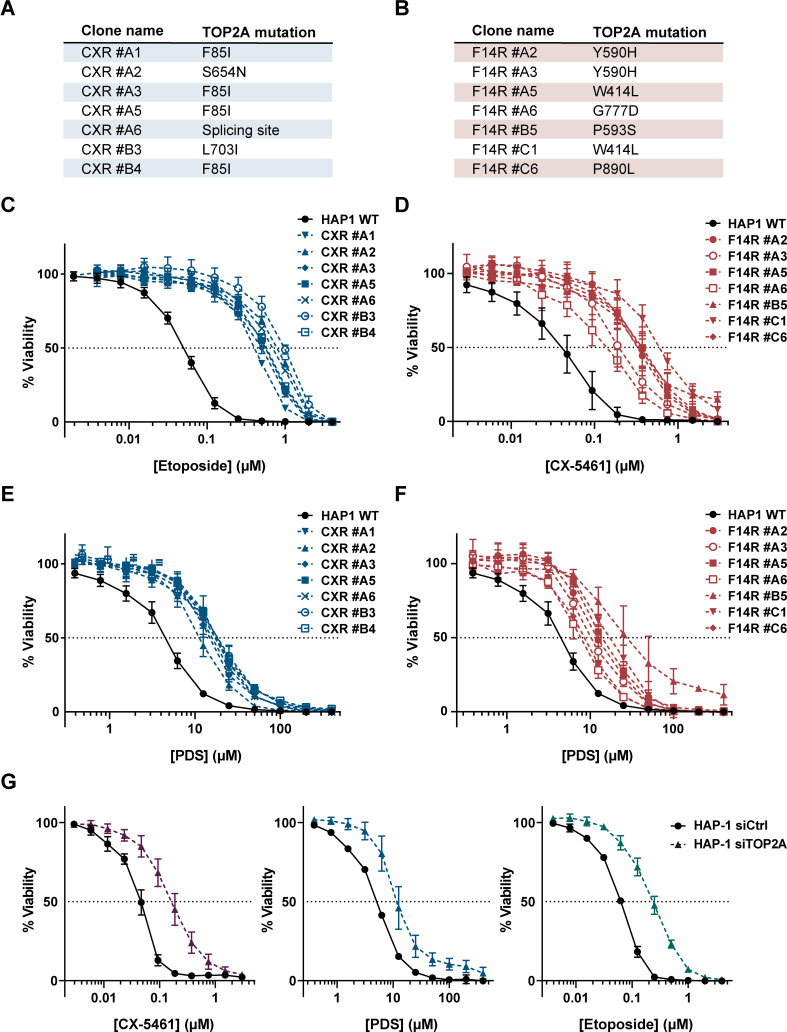
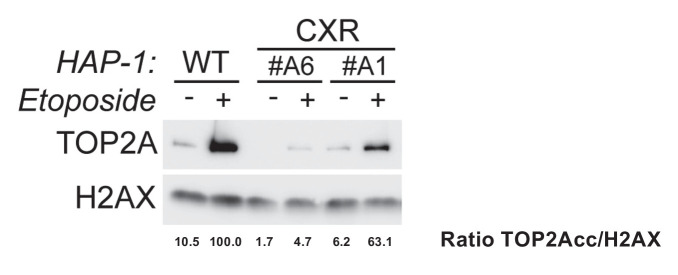
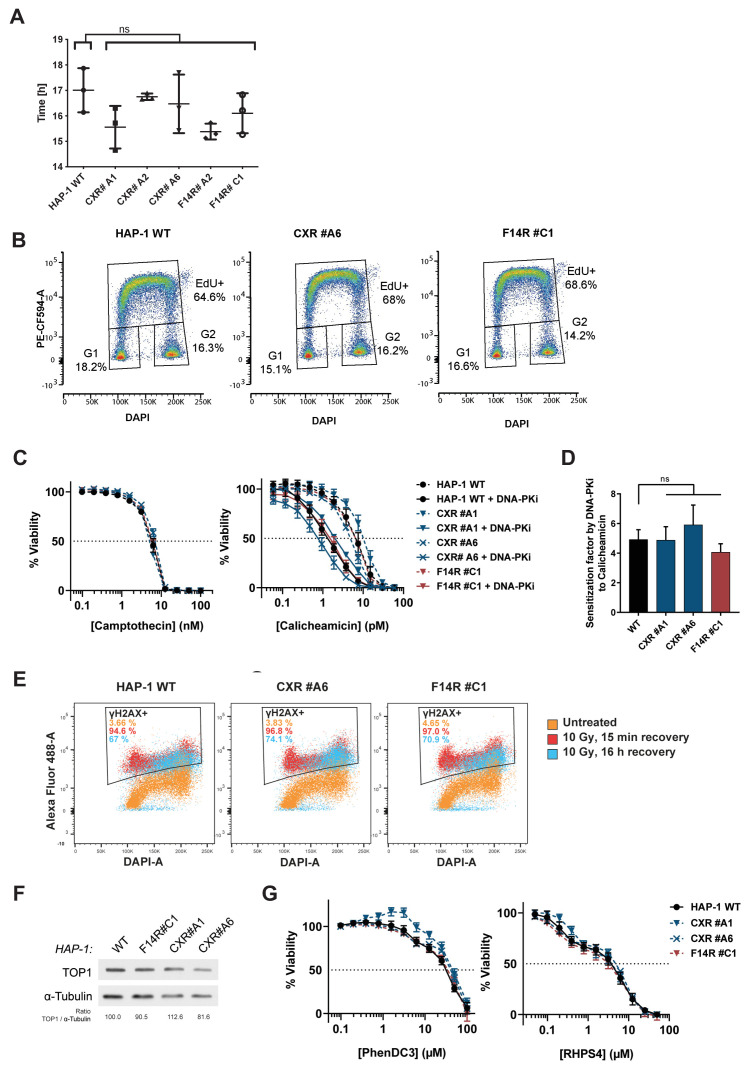
Figure 2. Impact of topoisomerase 2α (TOP2A) in the cell toxicity of topoisomerase poisons and G-quadruplex (G4) ligands.
(A, B) Summary of mutations found in CX-5461-resistant (CXR) (A) and F14R (B) clones. (C, E) Viability assay of CXR cells treated with etoposide (ETP) (C) and the G4 ligand pyridostatin (PDS) (E). (D, F) Viability assay of F14512-resistant cells (F14R) treated with the G4 ligands CX-5461 (D) and PDS (F). (G) Viability assay of TOP2A knock-down HAP1 cells treated with G4 ligand CX-5461 (left panel), PDS (central panel), and ETP (right panel). Error bars represent SD from the means, n ≥ 3 independent experiments.