Abstract
Objective
We developed and evaluated Drug-Drug Interaction Wide Association Study (DDIWAS). This novel method detects potential drug-drug interactions (DDIs) by leveraging data from the electronic health record (EHR) allergy list.
Materials and Methods
To identify potential DDIs, DDIWAS scans for drug pairs that are frequently documented together on the allergy list. Using deidentified medical records, we tested 616 drugs for potential DDIs with simvastatin (a common lipid-lowering drug) and amlodipine (a common blood-pressure lowering drug). We evaluated the performance to rediscover known DDIs using existing knowledge bases and domain expert review. To validate potential novel DDIs, we manually reviewed patient charts and searched the literature.
Results
DDIWAS replicated 34 known DDIs. The positive predictive value to detect known DDIs was 0.85 and 0.86 for simvastatin and amlodipine, respectively. DDIWAS also discovered potential novel interactions between simvastatin-hydrochlorothiazide, amlodipine-omeprazole, and amlodipine-valacyclovir. A software package to conduct DDIWAS is publicly available.
Conclusions
In this proof-of-concept study, we demonstrate the value of incorporating information mined from existing allergy lists to detect DDIs in a real-world clinical setting. Since allergy lists are routinely collected in EHRs, DDIWAS has the potential to detect and validate DDI signals across institutions.
Keywords: drug interactions, drug-related side effects, adverse reactions, pharmacovigilance, electronic health records, data mining
INTRODUCTION
Patients are taking more prescription drugs than ever to treat their chronic health conditions.1 This rise in drug use increases their risk of developing drug-drug interactions (DDIs).2 Patients experience DDIs when they concomitantly use an object drug (affected by the interaction) and a precipitant drug (causes the interaction). DDIs are responsible for > 20% of adverse drug reactions (ADRs)3 and for half of withdrawn drugs from the US market.4
DDIs can be recognized during drug development and clinical trials, but a lack of consensus for defining clinically actionable DDIs remains.5–8 Before a new drug is approved, potentially harmful DDIs are assessed using in vitro and in vivo methods. But it is not feasible to test for all the possible interactions between the new drug and those prescribed to patients.9 To identify DDIs missed during drug development, healthcare providers can voluntarily report DDIs to postmarket surveillance programs.10,11 Yet, underreporting of DDI events can occur, as DDIs are hard to recognize and reporting events may not be the highest priority for healthcare providers. To complement postmarket surveillance programs, researchers have developed methods to mine electronic health record (EHR) data for DDIs.12,13 Implementing these methods across EHRs, however, remains challenging, because they are either purpose-built14 or depend upon complex natural language processing (NLP).12
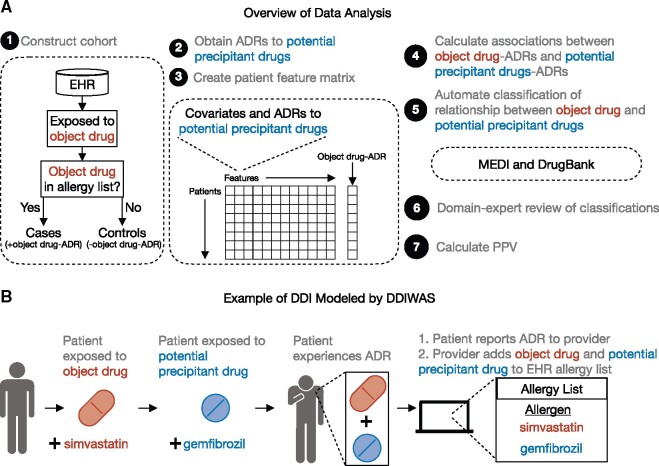
We developed Drug-Drug Interaction Wide Association Study (DDIWAS), a novel framework to identify potentially harmful DDIs by leveraging the EHR allergy list (Figure 1A). The allergy list is used by healthcare providers to document immune-mediated allergic drug reactions (eg, penicillin anaphylaxis15) and drug intolerances (eg, statin myopathy16) (Figure 1B). Allergy list entries also routinely contain only 2 data elements, the allergen (eg, culprit drug’s name) and reaction (eg, “muscle cramp”).17 This standardized pattern shared among EHRs enables high-throughput DDI detection without sophisticated NLP. In this study, we assumed that a drug’s appearance on the allergy list indicated that a drug ADR occurred. With that assumption, we hypothesized that adversely interacting drugs would frequently be documented together on the allergy list. We only used allergy list data because EHR fragmentation can make it difficult to obtain accurate medication lists.18
Figure 1.
Overview of data analysis and example of DDIs modeled by DDIWAS. (A) From a cohort of object drug-exposed patients, cases were those who had the object drug listed in their EHR allergy lists (+object drug-ADR), and controls were those who did not have the object drug documented on their allergy lists (−object drug-ADR). In this study, DDIWAS was applied on 2 object drugs, simvastatin and amlodipine. To search for potential precipitant drugs that increased the risk of object drug-ADRs, a systematic association test was performed using logistic regression. Potential precipitant drugs of interest were those that were positively associated with object-drug ADRs (logistic regression Bonferroni P value < .05 and OR > 1). Using MEDI and DrugBank, the relationship between the object drug and potential precipitant drugs were determined. All object-potential precipitant drug relationships were manually reviewed by a domain expert (SN, a pharmacist). PPV was then used to evaluate DDIWAS’ ability to replicate known DDIs. See also Supplementary Figure 1. (B) In this example of a DDI modeled by DDIWAS, the object drug is simvastatin, and the potential precipitant drug is gemfibrozil. The patient develops an ADR after concurrently using simvastatin and gemfibrozil. At the next visit, the patient reports their ADR to their provider who adds both drugs to the patient’s allergy list.
Abbreviations: ADR, adverse drug reaction; DDI, drug-drug interaction; DDIWAS, Drug-Drug Interaction Wide Association Study; EHR, electronic health record; MEDI, MEDication Indication resource; OR, odds ratio; PPV, positive predictive value.
To start the DDIWAS pipeline, we first identified a cohort of object drug-exposed patients in a deidentified EHR database19 (Figure 1A;Supplementary Figure 1). We then divided the patients into cases (+object drug ADR, ie, object drug on allergy list) and controls (−object drug ADR, ie, object drug not on allergy list). We searched for potential precipitant drugs that were disproportionately codocumented with the object drug on patients’ allergy lists. To measure DDIWAS performance, we calculated a positive predictive value (PPV) using a gold standard reference comprised of MEDication Indication resource (MEDI),20 DrugBank,21 and domain expert review (Supplementary Figure 2). We validated DDIWAS by applying it on 2 common drugs, simvastatin and amlodipine.
MATERIALS AND METHODS
Defining a drug-drug interaction (DDI)
In this study, a patient has experienced a DDI when the pharmacologic effects of 2 drugs overlap to produce an adverse outcome. When the object and precipitant drug interact, the patient experiences an ADR. The patient reports the ADR to their healthcare provider, who documents the adverse reaction in the patient’s EHR by adding the object drug to the patient’s allergy list. The provider does so because they believe that the ADR was most likely related to the patient’s exposure to the object drug.22 If the provider believes that the ADR was due to a DDI between the object and precipitant drug, then they may add both drugs to the patient’s allergy list.
As a concrete example of how DDIWAS determines whether a potential DDI occurred using allergy list data, consider a DDI between simvastatin (the object drug) and gemfibrozil (the precipitant drug) (Figure 1B). A provider prescribes gemfibrozil to a patient already on simvastatin. At the subsequent visit, the provider learns that after starting gemfibrozil, the patient began experiencing muscle aches. Since muscle ache is a common ADR associated with simvastatin exposure,23 the provider believes that a DDI between simvastatin and gemfibrozil occurred and adds both drugs to the patient’s allergy list.
Study design
The study was reviewed and approved by the IRB at Vanderbilt University Medical Center (VUMC) (#180456). We used deidentified EHR data from VUMC. The EHR database maintains longitudinal clinical data for over 3.2 million unique patients from inpatient and outpatient encounters.19 EHR data commonly includes diagnosis and procedure codes, medications, laboratory test results, unstructured clinical text, and demographics. We used EHR data from outpatient visits from 1996–2020 and limited our analyses to adult patients between the ages of 18 and 90 years.
To demonstrate the feasibility of DDIWAS, we used it to identify DDIs for simvastatin and amlodipine, drugs that are commonly used with known precipitant drugs.24 Simvastatin is 1 of the first-line therapies for hyperlipidemia and has a relatively increased frequency of myopathy at high doses.25 Amlodipine is commonly used to treat hypertension and is known to inhibit CYP3A4,23 a key enzyme involved in drug metabolism.
To identify drugs in the EHR, we used a standard terminology that formalizes all prescription drugs currently marketed in the US, RxNorm.26 We used generic and brand names to first map drugs to RxNorm Concept Unique Identifiers (RxCUIs) and then to their respective drug ingredients, based on their relationships in RxNorm. For example, “Simvastatin” (RxCUI 36567) and “Zocor” (RxCUI 196503) were both mapped to the drug ingredient “simvastatin” (RxCUI 36567).
For each object drug, we started with a cohort of patients who had ≥1 exposure(s) to the object drug (Figure 1A;Supplementary Figure 1A). In this cohort, we defined cases as patients who had the object drug documented on their allergy lists (+object drug-ADR), and defined controls as patients who did not have the object drug listed on their allergy lists (−object drug-ADR).
For both cases and controls, we set the date on which object drug exposure occurred as the start of the observation period (T0) (Supplementary Figure 1B). For cases, we set the date on which the object drug was first documented on their allergy list as the end of the observation period (Te). We limited the duration of the observation period to 12 months, because we wanted to capture ADRs from both short and long object drug exposures.27 If the observation period (Te–T0) was longer than 12 months, then we limited our analysis to the 12-month period prior to Te. For controls, we set Te as the date on which object drug exposure was last documented in their EHRs. If the observation period was longer than 12 months, we limited our analysis to the 12-month period after T0.
We obtained potential precipitant drug-ADRs by extracting all drugs documented on the patient allergy lists during the observation period (Supplementary Figure 1C). We then mapped the potential precipitant drugs to their RxCUI ingredients (Supplementary Figure 1D). To obtain only ADRs potentially due to DDIs between object and potential precipitant drugs, we removed drugs that were present on patient allergy lists prior to the start of the observation period. To prevent false-positive associations due to the absence of allergy list entries, we excluded controls who did not have any allergy list entries during the observation period (Supplementary Figure 1E).
Data preprocessing and association analysis
We created a patient feature matrix with each row representing 1 patient and with columns representing features (Supplementary Figure 1F). Features included covariates and potential precipitant drug-ADRs. The covariates were age, sex, race, duration of observation period, and number of unique drug ingredient exposures during the observation period. We encoded potential precipitant drug-ADRs as dichotomous variables. We then only tested potential precipitant drugs for which the 2x2 contingency table had ≥1 patient in each cell (Supplementary Figure 1G) because our goal was to identify drugs that increased the likelihood of object drug-ADRs.
To identify potential precipitant drugs that increased the risk of object drug-ADRs, we used the patient feature matrix to perform a systematic association study with logistic regression (Supplementary Figure 1H). For each patient, the dependent variable indicated whether the patient was a case (+object drug-ADR; object drug on allergy list) or control (−object drug-ADR; object drug not on allergy list). For each potential precipitant drug tested, the dichotomous independent variable indicated whether the drug was listed on each patient’s allergy list. The logistic regression analysis was adjusted for the covariates described above. The outputs of the logistic regression analysis were association odds ratios (ORs) and P values. Due to the limitations of logistic regression with rare events,28 we used Firth regression for potential precipitant drugs that had <5 patients in each cell of the 2x2 contingency table. To account for multiple testing, we applied a Bonferroni correction with type I error rate set to 0.05. We considered a potential precipitant drug to have increased the risk of object drug-ADRs if the following conditions were met: (1) regression OR > 1 and (2) regression Bonferroni-corrected P value < .05 (Supplementary Figure 2). Drugs that met these conditions indicated that patients with the potential precipitant drug listed on their allergy lists (+potential precipitant drug-ADR) were more likely to have the object drug listed as well (+object drug-ADR).
Labeling of DDIWAS output
We labeled the potential precipitant drugs meeting the 2 conditions to help us interpret DDIWAS results and to measure the method’s ability to replicate known DDIs. We used 3 labels: “Exclude,” “True-positive,” and “False-positive” (Supplementary Figure 2). To automatically label these drugs, we leveraged MEDI20 and DrugBank21 resources. First, using MEDI and manual engineering, we tagged drugs as “Exclude” if they shared indications with the object drug. Indications were represented by the International Classification of Diseases Ninth Revision, Clinical Modification (ICD-9-CM) code(s)29 most specific for the object drug. As an example, for the simvastatin experiment, we used the hyperlipidemia diagnosis codes, ICD-9-CM 272.4 “Other and unspecified hyperlipidemia” and ICD-9-CM 272.2 “Mixed hyperlipidemia.”
Our gold standard reference for “True-positive” findings was DrugBank21 followed by domain expert review. We used DrugBank to identify potential precipitant drugs that were known to interact adversely with the object drug of interest. We labeled potential precipitant drugs as “True-positive” if the DrugBank description indicated that for either the object or potential precipitant drug, drug metabolism decreased, serum concentration increased, drug absorption increased, drug elimination decreased, or concurrent use increased the risk of ADRs (eg, rhabdomyolysis with simvastatin use). We then tagged the remaining unlabeled potential precipitant drugs as “False-positive.” For final classification of object and potential precipitant drug pairs, all labels were manually reviewed by a domain expert (SN, a pharmacist).
Measuring DDIWAS performance to replicate known DDIs
To quantify the performance of DDIWAS to replicate known DDIs, we used PPV (Supplementary Figure 2). PPV was calculated by dividing the number of “True-positive” drugs by the sum of “True-positive” and “False-positive” drugs. PPV represented the fraction of remaining drugs with previously reported DDIs with the object drug of interest.
Adjusting for potential confounders and sensitivity analysis
To identify associations that may have been confounded by indication(s) for each significantly associated potential precipitant drug, we independently adjusted the regression for indications represented by phecodes.30 To select indications for each potential precipitant drug, we used the 2 ICD-9-CM codes with the highest prevalence from MEDI.20 We mapped these ICD-9-CM codes to their respective phecodes using the ICD-9-CM to phecode v1.2 map. We then rolled up the mapped phecodes to their parent phecodes. Using the indication for digoxin as an example, ICD-9-CM 427.31 “Atrial fibrillation” phecode 427.21 “Atrial fibrillation” phecode 427 “Cardiac dysrhythmias.” To obtain phecode indications for ICD-10-CM codes in our study cohort, we used the ICD-10-CM to phecode v1.2 (beta) map.31
We also conducted a sensitivity analysis by calculating PPVs and true-positive counts using minimum patient count thresholds of 1, 5, 10, and 20 patients in each cell of the 2x2 contingency table (Supplementary Figure 3).
Data visualization
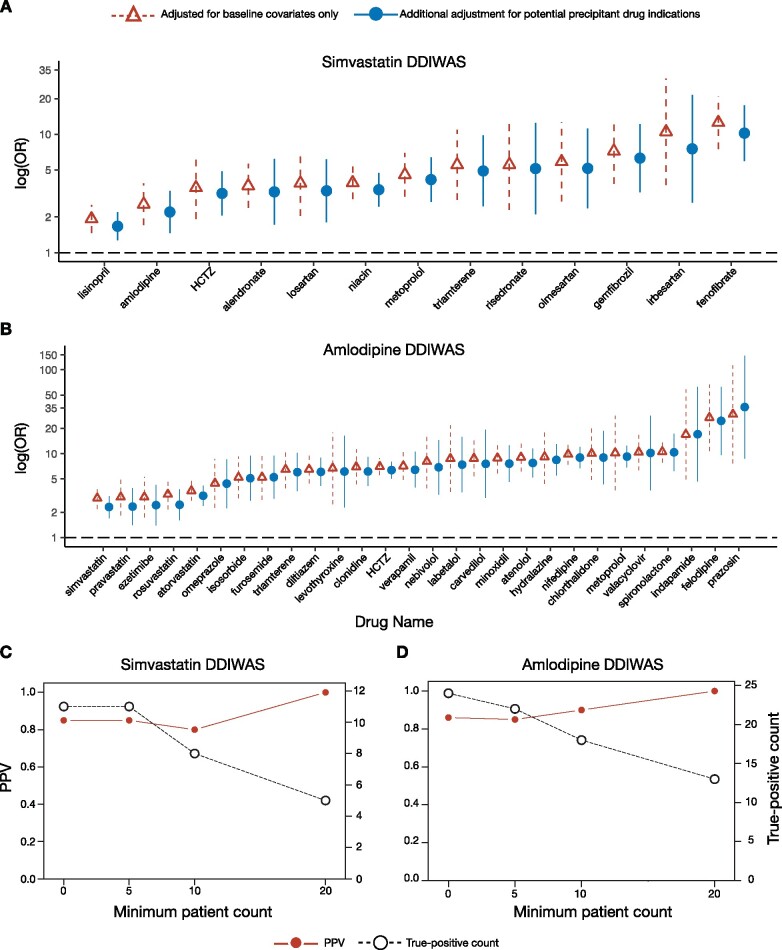
To present the results from our primary analysis, we used forest plots of regression OR (95% confidence interval [95% CI]) for all potential precipitant drugs that passed Bonferroni correction and OR > 1. For each drug, we show the OR (95% CI) from the logistic regression adjusted for baseline characteristics and a second regression with additional adjustment for each drug’s main indication(s) (Figures 2A and 2B).
Figure 2.
Forest plot of potential precipitant drugs associated with object drug ADRs and DDIWAS performance. (A, B) Forest plots summarizing the potential precipitant drugs that were significantly associated (logistic regression Bonferroni P value < .05 and OR > 1) with (A) simvastatin- and (B) amlodipine-ADRs. On the horizontal axis, potential precipitant drugs are sorted from smallest to largest ORs. On the vertical axis, association ORs (95% CI) are plotted on a logarithmic scale. Red triangles with dashed lines represent values from logistic regressions adjusted for age, sex, race, length of observation period, and number of unique drug exposures for each patient. Blue circles with solid lines indicate values from logistic regressions with additional adjustment for potential precipitant drug indications. These values were from analyses with a minimum patient count threshold of 1. Minimum patient count threshold refers to the number of patients required in each cell of the 2x2 contingency table (Supplementary Figure 3). The Bonferroni correction was 1.77x10−4 (0.05/282) for simvastatin and 1.49 x 10−4 (0.05/335) for amlodipine. See Supplementary Table 2 for corresponding numbers. (C, D) PPV (left vertical axis) and true-positive count (right vertical axis) for (C) simvastatin and (D) amlodipine DDIWAS at minimum patient count thresholds of 1, 5, 10, and 20. True-positive count refers to the number of potential precipitant drugs (logistic regression Bonferroni P value < .05 and OR > 1) that were known to interact with the object drug. See Supplementary Table 3 for corresponding numbers.
Abbreviations: ADR, adverse drug reaction; DDIWAS, Drug-Drug Interaction Wide Association Study; HCTZ, hydrochlorothiazide; PPV, positive predictive value; OR, odds ratio; 95% CI: 95% confidence interval.
Validation studies for potentially novel DDIs
We defined “False-positive” drugs as potentially novel DDIs (Supplementary Figure 2). To validate these potentially novel DDIs, we reviewed the clinical notes for 10 randomly selected patients who DDIWAS labeled as (+object drug-ADR, +potential precipitant drug-ADR). If there were less than 10 (+object drug-ADR, +potential precipitant drug-ADR) patients, we reviewed the clinical notes for all available patients. For each DDIWAS-labeled +drug-ADR patient, we reviewed their clinical notes to verify that the drug was intentionally added to their allergy lists. Each reviewed DDIWAS-labeled +drug-ADR patient was labeled “True-positive +drug-ADR” or “False-positive +drug-ADR.” A “True-positive +drug-ADR” patient was not exposed to the drug after the end of the observation period and/or for whom a provider mentioned the drug-ADR in additional EHR sections like “History of Present Illness” and “Assessment & Plan.” A “False-positive +drug-ADR” patient did not meet either criteria.
After reviewing patient charts, we were concerned about the remaining drugs that met our criteria for potentially novel DDIs, but whose associations with object drug-ADRs did not likely represent novel DDIs. Instead, the associations were more likely to be due to interactions between the object drug and other drugs; the other drugs were either commonly coprescribed or combined with the drugs of concern. To address this problem, we adjusted the regressions for additional potential precipitant drug-ADRs. For example, in the primary simvastatin DDIWAS analysis, the regression for triamterene was:
| (1) |
We then adjusted the regression for hydrochlorothiazide (HCTZ)-ADRs because HCTZ is frequently combined with triamterene:
| (2) |
RESULTS
Simvastatin DDIWAS
The simvastatin experiment (Table 1) had 85 873 controls (+simvastatin-exposed, -simvastatin-ADR) and 2814 cases (+simvastatin-exposed, +simvastatin-ADR). Of the 282 potential precipitant drugs tested (Supplementary Figure 4A; Supplementary Table 1), 13 increased the risk of simvastatin-ADRs (passing Bonferroni correction [0.05/282 = 1.77x10−4] with OR > 1; Figure 2A). To control for potential confounding by drug indications, we adjusted the regressions for potential precipitant drug indications and found that all 13 associations remained significant (Supplementary Table 2). Eleven of the 13 drugs were known to interact with simvastatin, including fenofibrate, gemfibrozil, niacin, and amlodipine. In DrugBank, the remaining 2 drugs not known to interact with simvastatin were HCTZ and triamterene.
Table 1.
Simvastatin patient-level characteristics
| Characteristic | Controls (n = 85 873) | Cases (n = 2814) | P Value |
|---|---|---|---|
| Female | 0.52 (44 367) | 0.56 (1564) | <.001 |
| White | 0.80 (68 900) | 0.85 (2384) | <.001 |
| Age, years | 63 (54–71) | 63 (54–70) | .15 |
| Observation period length, days | 337 (15–365) | 365 (111–365) | <.001 |
| Unique drug exposures, count | 12 (7–19) | 13 (8–25) | <.001 |
| Phecode 250.* (Diabetes mellitus) | 0.19 (16 241) | 0.25 (702) | <.001 |
| Phecode 272.* (Disorders of lipid metabolism) | 0.34 (28 776) | 0.74 (2,084) | <.001 |
| Phecode 401.* (Hypertensive disorder) | 0.35 (30 253) | 0.59 (1654) | <.001 |
| Phecode 411.* (Myocardial infarction) | 0.20 (17 384) | 0.31 (859) | <.001 |
| Phecode 418.* (Chest pain) | 0.13 (10 774) | 0.18 (519) | <.001 |
| Phecode 743.* (Osteoporosis) | 0.04 (3280) | 0.06 (172) | <.001 |
For continuous variables, numbers represent median (interquartile range).
For dichotomous variables, numbers after proportions are counts.
P values indicate differences between cases and controls. For continuous variables, P values were calculated using Mann-Whitney test. For dichotomous variables, P values were calculated using test. P < .05 was considered statistically significant.
Note: For phecodes, * means ≥1 digits or a period (eg, phecode 401.* = phecodes 401, 401.1, 401.2, 401.21, 401.22, or 401.3).
To examine the potential novel DDIs between simvastatin-HCTZ and simvastatin-triamterene, we manually reviewed clinical notes to verify that the drugs were intentionally listed on patient allergy lists (Table 2). The reviewed notes were from 2 types of patients: those who potentially experienced simvastatin-HCTZ DDIs, ie, DDIWAS-labeled (+simvastatin-ADR, +HCTZ-ADR) and those who potentially experienced simvastatin-triamterene DDIs, ie, DDIWAS-labeled (+simvastatin-ADR, +triamterene-ADR). All reviewed patients had the respective drugs intentionally listed on their allergy lists. We hypothesized that the triamterene association was confounded by HCTZ-ADRs, because all reviewed DDIWAS-labeled (+simvastatin-ADR, +triamterene-ADR) patients were exposed via a HCTZ/triamterene combination drug. Further, there were DDIWAS-labeled (+simvastatin-ADR, +HCTZ-ADR) patients who did not have triamterene on their allergy lists. To test our hypothesis, we adjusted the triamterene-ADR regression with HCTZ-ADRs and found that triamterene’s association was no longer significant, while HCTZ’s association remained significant (Supplementary Table 4).
Table 2.
Validation analysis of potentially novel DDIs, manual chart review results
| Object Drug | Potential Precipitant Drug | % TP Drug-ADR(TP/number of patients reviewed) | Comments |
|---|---|---|---|
| simvastatin | HCTZ | 100 (10/10) | NA |
| simvastatin | triamterene | 100 (10/10) | All DDIWAS-derived (+simvastatin-ADR, +triamterene-ADR) patients were exposed to triamterene via a HCTZ/triamterene combination drug. |
| amlodipine | ezetimibe | 90 (9/10) |
|
| amlodipine | levothyroxine | 40 (2/5) |
|
| amlodipine | valacyclovir | 80 (4/5) |
|
| amlodipine | omeprazole | 100 (10/10) | NA |
True-positive patients were those for whom healthcare providers intentionally added both the object and potential precipitant drugs to their allergy lists.
Abbreviations: ADR, adverse drug reaction; DDIWAS, Drug-Drug-Interaction Wide Association Study; HCTZ, hydrochlorothiazide; TP, true-positive.
Amlodipine DDIWAS
The amlodipine experiment (Table 3) had 83 732 controls (+amlodipine-exposed, -amlodipine-ADR) and 2512 cases (+amlodipine-exposed, +amlodipine-ADR). Of the 335 potential precipitant drugs tested (Supplementary Figure 4B; Supplementary Table 1), 28 increased the risk of amlodipine-ADRs (passing Bonferroni correction [0.05/335 = 1.49x10−4] with OR > 1; Figure 2B). All associations remained significant after adjusting the regressions for potential precipitant drug indications (Supplementary Table 2). Twenty-four of the 28 drugs were known to interact with amlodipine, including prazosin, diltiazem, and verapamil. In DrugBank, there were 4 drugs not known to interact with amlodipine: levothyroxine, ezetimibe, omeprazole, and valacyclovir.
Table 3.
Amlodipine patient-level characteristics
| Characteristic | Controls (n = 83 732) | Cases (n = 2512) | P |
|---|---|---|---|
| Female | 0.54 (45 315) | 0.65 (1637) | <.001 |
| White | 0.75 (63 144) | 0.83 (2083) | <.001 |
| Age, years | 63 (53–72) | 65 (55–73) | <.001 |
| Observation period length, days | 287 (14–365) | 206 (38–365) | .91 |
| Unique drug exposures, count | 12 (8–20) | 13 (8–22) | <.001 |
| Phecodes 053.* (Herpes zoster) | 2.93E-03 (245) | 3.18E-03 (8) | .81 |
| Phecodes 054.* (Herpes simplex) | 1.97E-03 (165) | 2.39E-03 (6) | .64 |
| Phecodes 244.* (Hypothyroidism) | 0.05 (4558) | 0.08 (201) | <.001 |
| Phecodes 250.* (Diabetes mellitus) | 0.16 (13 144) | 0.15 (365) | .11 |
| Phecodes 272.* (Disorders of lipid metabolism) | 0.21 (17 898) | 0.37 (927) | <.001 |
| Phecodes 300.* (Anxiety, phobic and dissociative disorders) | 0.04 (3325) | 0.05 (114) | .15 |
| Phecodes 401.* (Hypertensive disorder) | 0.43 (36 059) | 0.68 (1702) | <.001 |
| Phecodes 411.* (Myocardial infarction) | 0.14 (11 898) | 0.15 (371) | .43 |
| Phecodes 414.* (Other forms of chronic heart disease) | 0.02 (1968) | 0.02 (46) | .09 |
| Phecodes 418.* (Chest pain) | 0.11 (9170) | 0.14 (348) | <.001 |
| Phecodes 427.* (Cardiac dysrhythmias) | 0.12 (10 096) | 0.14 (355) | .002 |
| Phecodes 428.* (Congestive heart failure) | 0.06 (4609) | 0.05 (138) | .98 |
| Phecodes 530.* (Esophageal disorders) | 0.08 (6706) | 0.09 (228) | .053 |
| Phecodes 536.* (Disorders of function of stomach) | 0.01 (858) | 0.01 (18) | .13 |
For continuous variables, numbers represent median (interquartile range).
For dichotomous variables, numbers after proportions are counts.
P values indicate differences between cases and controls. For continuous variables, P values were calculated using Mann-Whitney test. For dichotomous variables, P values were calculated using test. P < .05 was considered statistically significant.
For phecodes, * means ≥1 digit or a period (eg, phecode 401.* = phecodes 401, 401.1, 401.2, 401.21, 401.22, or 401.3).
To examine the potential novel DDIs between amlodipine and the 4 drugs, we manually reviewed clinical notes (Table 2). First, for levothyroxine, of the 5 available DDIWAS-labeled (+amlodipine-ADR, +levothyroxine-ADR) patients, 2 had both drugs listed on their allergy lists, 1 had only amlodipine listed, and 2 had neither drug listed. Of note, the 2 false-positive DDIWAS-labeled (+amlodipine-ADR, +levothyroxine-ADR) patients were taking both drugs during the observation period. Second, for ezetimibe, 90% (9/10) of DDIWAS-labeled (+amlodipine-ADR, +ezetimibe-ADR) patients had both drugs on their allergy lists. The single false-positive DDIWAS-labeled (+amlodipine-ADR, +ezetimibe-ADR) patient did not have either drug listed on their allergy list. Since ezetimibe is commonly used with statins to lower cholesterol, we then adjusted the ezetimibe regression for ADRs to common statins, simvastatin and atorvastatin; in this statin-ADRs adjusted regression, the association P value for ezetimibe was no longer significant (P value = .29; Supplementary Table 4) However, in this same adjusted ezetimibe regression, the association P values for both statin-ADRs remained significant. Third, for omeprazole, all (10/10) DDIWAS-labeled (+amlodipine-ADR, +omeprazole-ADR) patients had both drugs documented on their allergy lists. Fourth, for valacyclovir, 4/5 DDIWAS-labeled (+amlodipine-ADR, +valacyclovir-ADR) patients had both drugs documented on their allergy lists.
Replication sensitivity analysis
To quantify the performance of DDIWAS to replicate known DDIs, we calculated the PPV for both simvastatin and amlodipine experiments at minimum patient count thresholds of 1, 5, 10, and 20 (Supplementary Table 3). In the simvastatin experiment, as thresholds increased, the PPV increased from 0.85 to 1.00, but the number of true-positive findings decreased from 11 to 5 potential precipitant drugs (Figure 2C). For amlodipine, as thresholds increased, the PPV increased from 0.86 to 1.00, but the number of true-positive findings decreased from 24 to 13 (Figure 2D).
DISCUSSION
DDIWAS is a high-throughput method to identify potential DDIs by mining the EHR allergy list. We used the method to identify potential DDIs for simvastatin and amlodipine. DDIWAS replicated known DDIs with a PPV of 0.85 and 0.86 for simvastatin and amlodipine, respectively. For both drugs, DDIWAS also detected potentially novel DDIs that were validated with manual review of patient clinical notes. Our validation studies support potentially novel interactions between simvastatin-HCTZ, amlodipine-omeprazole, and amlodipine-valacyclovir.
Existing methods to mine EHR data have successfully replicated known DDIs,12 but have limitations that prevent widespread adoption. First, the tools used to detect DDIs in EHRs are rarely publicly available. Second, even if they are available, these tools are often purpose-built advanced NLP or text annotation applications,32 requiring users to perform substantial customization for use with external datasets.33,34 In contrast, DDIWAS identifies DDI events using drug name recognition, a relatively simpler task than NLP-based detection of ADRs. Recognizing drug names is easier than detecting ADRs across health systems due to local documentation procedures that may lead to differences in how ADRs are represented in clinical narratives.35,36 DDIWAS may be easier to implement in external databases, as it only searches for drug names in medication and allergy lists and EHR modules with smaller contextual variability than in clinical narratives. We anticipate that users will be able to apply DDIWAS to identify DDIs in their databases, without spending substantial time and resources to modify text annotation tools.
To test our approach to identify DDIs, we wanted to see whether we could replicate drugs known to interact with the object drugs, simvastatin and amlodipine. We found that 85% (35/41) of the significantly associated drugs (Bonferroni P value < .05 and OR > 1) were known to interact with the object drugs. In the simvastatin analysis, we tested 8 drugs that were recommended for inclusion in all clinical decision support (CDS) DDI alert systems.37 These drugs were amiodarone, clarithromycin, diltiazem, erythromycin, fluconazole, ketoconazole, nefazodone, and verapamil. Among these drugs, none were found to be significantly associated with simvastatin-ADRs. These “false-negative” findings could partially be attributed to intervention by the CDS alerts designed to reduce cases of clinically significant DDIs.38,39 Notably, drugs that were significantly associated with simvastatin-ADRs included niacin and warfarin. Although these drugs are known to interact adversely with simvastatin, an expert committee recommended that alerts for these DDIs be deleted because the therapeutic benefits of these drugs outweigh the risk of patient harm.40 In this study’s amlodipine analysis, prazosin’s association had the largest effect size (Figure 2B;Supplementary Table 1). This finding is supported in the literature, as patients using both calcium-channel blockers (eg, amlodipine) and alpha-1 blockers (eg, prazosin) have been found to be at increased risk of developing hypotension.41,42
In addition to replicating known DDIs, DDIWAS also identified potentially novel DDIs. Our results suggest a potential novel simvastatin-HCTZ DDI. Out of the 13 drugs significantly associated with simvastatin-ADRs, HCTZ and triamterene did not have previously reported DDIs with simvastatin. When the triamterene regression was adjusted for HCTZ-ADRs, HCTZ’s association, but not triamterene’s, was still significant at a Bonferroni P value < .05 (Supplementary Table 4). It has been shown that patients who concurrently used statins and HCTZ were at increased risk of adverse events, including chest pain, hyperglycemia, and muscle spasms.43 Additional evidence to support a simvastatin-HCTZ DDI can be found in DrugBank; rosuvastatin and pravastatin are predicted to decrease HCTZ excretion, suggesting a possible interaction between HCTZ and the statin drug class. Nonetheless, a biological mechanism to explain a simvastatin-HCTZ interaction remains to be explored.
DDIWAS found potentially novel amlodipine-DDIs with valacyclovir and omeprazole. A previous study has shown that patients exposed concurrently to amlodipine and valacyclovir were at increased risk of developing adverse outcomes like acute kidney failure, dysarthria, and dizziness.43 The same study found that patients using both amlodipine and omeprazole were more likely to experience chest pain and dyspnea.43 A pharmacogenomic study found that CYP2C19 intermediate metabolizers were more prone to developing amlodipine-omeprazole DDIs.44 When exposed to both amlodipine and omeprazole, these patients experienced higher than expected drops in blood pressure. The authors proposed a mechanism in which elevated levels of omeprazole inhibits CYP3A metabolism of amlodipine, leading to lower blood pressure. Overall, results from the amlodipine experiments corroborate DDIWAS as an effective tool to detect potentially novel DDIs using real-world evidence in EHR data.
There are several limitations in this study. First, to detect DDIs, DDIWAS uses frequentist approaches assuming no prior information. If there is prior knowledge of a DDI, such as those derived from pharmacologic and/or pharmacokinetic studies, we can potentially improve DDIWAS using Bayesian approaches with prior probabilities determined from existing evidence.45,46 Second, we only performed DDIWAS using a maximum observation window length of 1 year and did not examine other period lengths. Third, we assumed that a patient experienced an adverse outcome to a drug of interest if the drug was listed on the patient’s allergy list. Even if a healthcare provider intentionally added a drug to a patient’s allergy list, the patient still may not have truly experienced an ADR to the drug. Potential reasons for false-positive cases include unverified patient-reported ADRs,47,48 disease exacerbation presenting like an ADR, and variability among healthcare providers’ abilities to identify the causal drug.49 But multiple studies have successfully used the allergy list to identify patients with ADRs.50,51 Likewise, we found that the majority of the DDIWAS-labeled +drug-ADR patients reviewed truly had the drugs listed on their allergy lists (Table 2). The dependence on healthcare providers’ abilities to correctly identify causal drugs also increases the probability of false-negative DDIs. For example, DDIWAS did not detect a well-known interaction between simvastatin and amiodarone.16 It would be interesting to see whether using drug exposures from the medication list increases the sensitivity of DDIWAS to identify potential DDIs without sacrificing PPV. Fourth, in its current form, DDIWAS does not systematically adjust for combination drugs, which can confound the interpretation of associations. In the simvastatin experiment, our stratified analysis found that the simvastatin-triamterene association was confounded by patients taking HCTZ/triamterene combination drugs (Table 2; Supplementary Table 4). Drugs frequently coprescribed can also contribute to false-positive findings. We found that the amlodipine-ezetimibe association was most likely confounded by interactions between amlodipine and statin drugs. A module to automatically adjust associations for combination drugs and drugs often used together is an opportunity for future development. Fifth, to maximize the transportability of DDIWAS, we did not use ADR information that was present in some allergy list entries. Using ADR information represented as unstructured text would likely require NLP expertise, as providers may describe the same ADRs differently (eg, myopathy could be described as “muscle cramp,” “myotoxicity,” “muscle weakness”). Recently, Wang et al developed a data-driven approach to help providers pick specific ADRs conditional on the drug selected in the allergy list.17 Incorporating such approaches may increase the use of structured ADR entries, which could augment DDIWAS’ ability to detect potential DDIs. Sixth, like other retrospective observational studies, we do not claim that these associations were caused by DDIs. Like previous studies,12 our goal was to show that DDIWAS can generate DDI hypotheses that will require validation by follow-up studies. Last, while we applied DDIWAS to data from only 1 institution, users at external institutions that also organize their EHR data with the Observational Health Data Sciences and Informatics (OHDSI)/Observational Medical Outcomes Partnership (OMOP) Common Data Model52 can apply DDIWAS to their dataset after making minor changes to the code that we have shared publicly.53
CONCLUSIONS
In summary, we developed and evaluated DDIWAS, a novel method that uses EHR allergy list entries to detect DDIs. DDIWAS replicated known DDIs and identified potentially novel DDIs. EHR-based methods like DDIWAS could complement existing tools to improve postmarket surveillance of DDIs.
FUNDING
This work was supported by National Institutes of Health grant numbers T32GM007347, R01 LM010685, R01 HL133786, T15 LM007450, P50 GM115305, K12HS026395, R35 GM131770, and American Heart Association grant number 16SDG27490014. The data set used for the analyses described was obtained from Vanderbilt University Medical Center’s resources, the Synthetic Derivative, which are supported by institutional funding and by the National Center for Advancing Translational Science grant number 2UL1 TR000445-06. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
AUTHOR CONTRIBUTIONS
Study initialization: PW, JCD, and W-QW; study design: PW, SDN, QF, QC, JCD, and W-QW; acquisition of data: PW and W-QW; analysis and interpretation of data: PW, SDN, JZ, CS, QF, QC, EAL, BL, NC, CMS, EJP, DMR, JCD, and W-QW; drafting of the manuscript: PW and W-QW.
All authors contributed to refinement of the manuscript and approved the final manuscript.
Grant holders: W-QW, EJP, DMR, and JCD.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank David Schlueter (National Institutes of Health) for his advice on statistical analysis. We would like to thank Raymond Wu, Aaron Lim (Vanderbilt University School of Medicine), Bryan Steitz (Vanderbilt University School of Medicine), and Vivian Siegel (Vanderbilt University Medical Center) for reading drafts of this manuscript.
DATA AVAILABILITY STATEMENT
Due to patient privacy concerns, we are unable to share the EHR data used in this study. However, we have released the DDIWAS R package on GitHub (https://github.com/pwatrick/ddiwas) under an Apache License and archived version 0.1 on Zenodo.53 We have written a tutorial to extract data from EHR databases organized using the OHDSI/OMOP Common Data Model,52 which can be found at https://pwatrick.github.io/ddiwas/articles/extract_ehr_data.html. We have also prepared a guide to process and analyze extracted EHR data, which can be found at https://pwatrick.github.io/ddiwas/articles/ddiwas_r_package_tutorial.html.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Kantor ED, Rehm CD, Haas JS, et al. Trends in prescription drug use among adults in the united states from 1999-2012. JAMA 2015; 314 (17): 1818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnell K, Klarin I.. The relationship between number of drugs and potential drug-drug interactions in the elderly: a study of over 600,000 elderly patients from the Swedish prescribed drug register. Drug Saf 2007; 30 (10): 911–8. [DOI] [PubMed] [Google Scholar]
- 3. Magro L, Moretti U, Leone R.. Epidemiology and characteristics of adverse drug reactions caused by drug-drug interactions. Expert Opin Drug Saf 2012; 11 (1): 83–94. [DOI] [PubMed] [Google Scholar]
- 4. Huang S-M, Lesko LJ.. Drug-drug, drug-dietary supplement, and drug-citrus fruit and other food interactions: What have we learned? J Clin Pharmacol 2004; 44 (6): 559–69. [DOI] [PubMed] [Google Scholar]
- 5. Olvey EL, Clauschee S, Malone DC.. Comparison of critical Drug–Drug interaction listings: The Department of Veterans Affairs medical system and standard reference compendia. Clin Pharmacol Ther 2010; 87 (1): 48–51. [DOI] [PubMed] [Google Scholar]
- 6. Phansalkar S, Desai A, Choksi A, et al. Criteria for assessing high-priority drug-drug interactions for clinical decision support in electronic health records. BMC Med Inform Decis Mak 2013; 13 (1): 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vitry AI. Comparative assessment of four drug interaction compendia. Br J Clin Pharmacol 2007; 63 (6): 709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fung KW, Kapusnik-Uner J, Cunningham J, et al. Comparison of three commercial knowledge bases for detection of drug-drug interactions in clinical decision support. J Am Med Inform Assoc 2017; 24 (4): 806–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang S-M, Temple R, Throckmorton DC, et al. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin Pharmacol Ther 2007; 81 (2): 298–304. [DOI] [PubMed] [Google Scholar]
- 10.Clinical Drug Interaction Studies—Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions Guidance for Industry. Rockville, MD: Food and Drug Administration Draft Guidance; 2017; US Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER). https://wayback.archive-it.org/7993/20190422131516/https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugs-gen/documents/document/ucm292362.pdf Accessed 6 March 6, 2021.
- 11. Center for Drug Evaluation, Research. Questions and Answers on FDA’s Adverse Event Reporting System (FAERS). US Food and Drug Administration; 2019. http://www.fda.gov/drugs/surveillance/questions-and-answers-fdas-adverse-event-reporting-system-faers Accessed October 9, 2019.
- 12. Iyer SV, Harpaz R, LePendu P, et al. Mining clinical text for signals of adverse drug-drug interactions. J Am Med Inform Assoc 2014; 21 (2): 353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011; 90 (1): 133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lorberbaum T, Sampson KJ, Chang JB, et al. Coupling data mining and laboratory experiments to discover drug interactions causing QT prolongation. J Am Coll Cardiol 2016; 68 (16): 1756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Warrington R, Silviu-Dan F.. Drug allergy. Allergy Asthma Clin Immunol 2011; 7 (Suppl 1): S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newman CB, Preiss D, Tobert JA, et al. Statin safety and associated adverse events: A scientific statement from the american heart association. Arterioscler Thromb Vasc Biol 2019; 39: e38–81. [DOI] [PubMed] [Google Scholar]
- 17. Wang L, Blackley SV, Blumenthal KG, et al. A dynamic reaction picklist for improving allergy reaction documentation in the electronic health record. J Am Med Inform Assoc 2020; 27: 917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei W-Q, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J Am Med Inform Assoc 2012; 19 (2): 219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84 (3): 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wei W-Q, Cronin RM, Xu H, et al. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc 2013; 20 (5): 954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res 2018; 46 (D1): D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McConeghy KW, Caffrey AR, Morrill HJ, et al. Are non-allergic drug reactions commonly documented as medication ‘allergies’? A national cohort of veterans’ admissions from 2000 to 2014. Pharmacoepidemiol Drug Saf 2017; 26 (4): 472–6. [DOI] [PubMed] [Google Scholar]
- 23. Wiggins BS, Saseen JJ, Page RL 2nd, et al. Recommendations for management of clinically significant Drug-Drug interactions with statins and select agents used in patients with cardiovascular disease: a scientific statement from the American Heart Association. Circulation 2016; 134 (21): e468–95. [DOI] [PubMed] [Google Scholar]
- 24. Qato DM, Wilder J, Schumm LP, et al. Changes in prescription and over-the-counter medication and dietary supplement use among older adults in the United States, 2005 vs 2011. JAMA Intern Med 2016; 176 (4): 473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Armitage J, Bowman L, Wallendszus K; SEARCH Collaborative Group, et al. Intensive lowering of LDL cholesterol with 80 mg versus 20 mg simvastatin daily in 12,064 survivors of myocardial infarction: a double-blind randomised trial. Lancet 2010; 376 (9753): 1658–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011; 18 (4): 441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Molokhia M, McKeigue P, Curcin V, et al. Statin induced myopathy and myalgia: Time trend analysis and comparison of risk associated with statin class from 1991-2006. PLoS One 2008; 3 (6): e2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heinze G, Schemper M.. A solution to the problem of separation in logistic regression. Statist Med 2002; 21 (16): 2409–19. [DOI] [PubMed] [Google Scholar]
- 29. Steindel SJ. International classification of diseases, 10th edition, clinical modification and procedure coding system: descriptive overview of the next generation HIPAA code sets. J Am Med Inform Assoc 2010; 17 (3): 274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013; 31 (12): 1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform 2019; 7 (4): e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lependu P, Iyer SV, Fairon C, et al. Annotation analysis for testing drug safety signals using unstructured clinical notes. J Biomed Semant 2012; 3 (S1): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc 2011; 18 (5): 540–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng K, Vydiswaran VGV, Liu Y, et al. Ease of adoption of clinical natural language processing software: an evaluation of five systems. J Biomed Inform 2015; 58: S189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harpaz R, Callahan A, Tamang S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf 2014; 37 (10): 777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosenbloom ST, Carroll RJ, Warner JL, et al. Representing knowledge consistently across health systems. Yearb Med Inform 2017; 26 (01): 139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phansalkar S, H van der S, Tucker AD, et al. Drug-drug interactions that should be non-interruptive in order to reduce alert fatigue in electronic health records. J Am Med Inform Assoc 2013; 20 (3): 489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sutton RT, Pincock D, Baumgart DC, et al. An overview of clinical decision support systems: benefits, risks, and strategies for success. NPJ Digit Med 2020; 3 (1): 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Helmons PJ, Suijkerbuijk BO, Nannan Panday PV, et al. Drug-drug interaction checking assisted by clinical decision support: a return on investment analysis. J Am Med Inform Assoc 2015; 22 (4): 764–72. [DOI] [PubMed] [Google Scholar]
- 40. Phansalkar S, Desai AA, Bell D, et al. High-priority drug-drug interactions for use in electronic health records. J Am Med Inform Assoc 2012; 19 (5): 735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elliott HL, Meredith PA, Campbell L, et al. The combination of prazosin and verapamil in the treatment of essential hypertension. Clin Pharmacol Ther 1988; 43 (5): 554–60. [DOI] [PubMed] [Google Scholar]
- 42. Lenz ML, Pool JL, Laddu AR, et al. Combined terazosin and verapamil therapy in essential hypertension. hemodynamic and pharmacokinetic interactions. Am J Hypertens 1995; 8 (2): 133–45. [DOI] [PubMed] [Google Scholar]
- 43. Tatonetti NP, Ye PP, Daneshjou R, et al. Data-driven prediction of drug effects and interactions. Sci Transl Med 2012; 4 (125): 125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dorofeeva MN, Shikh EV, Sizova ZM, et al. Antihypertensive effect of amlodipine in co-administration with omeprazole in patients with hypertension and acid-related disorders: cytochrome p450-associated aspects. PGPM 2019;12: 329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Almenoff JS, DuMouchel W, Kindman LA, et al. Disproportionality analysis using empirical bayes data mining: a tool for the evaluation of drug interactions in the postmarketing setting. Pharmacoepidem Drug Safe 2003; 12 (6): 517–21. [DOI] [PubMed] [Google Scholar]
- 46. E van de S, Venhorst J, Jansen HT, et al. Generation of bayesian prediction models for OATP-mediated drug-drug interactions based on inhibition screen of OATP1B1, OATP1B1∗15, and OATP1B3. Eur J Pharm Sci 2015; 70: 29–36. [DOI] [PubMed] [Google Scholar]
- 47. Trubiano JA, Adkinson NF, Phillips EJ.. Penicillin allergy is not necessarily forever. JAMA 2017; 318 (1): 82–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stone CA Jr, Trubiano J, Coleman DT, et al. The challenge of de-labeling penicillin allergy. Allergy May 2019; 75 (2): 273–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grizzle AJ, Hines LE, Malone DC, et al. Testing the face validity and inter-rater agreement of a simple approach to drug-drug interaction evidence assessment. J Biomed Inform 2020; 101: 103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krebs K, Bovijn J, Zheng N, et al. Genome-wide study identifies association between HLA-B∗55:01 and self-reported penicillin allergy. Am J Hum Genet 2020; 107: 612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiley LK, Moretz JD, Denny JC, et al. Phenotyping adverse drug reactions: statin-related myotoxicity. AMIA Jt Summits Transl Sci Proc 2015; 2015: 466–70. [PMC free article] [PubMed] [Google Scholar]
- 52. Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012; 19 (1): 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu P. pwatrick/ddiwas: First commit. Zenodo2020. 10.5281/zenodo.4251662 Accessed November 6, 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to patient privacy concerns, we are unable to share the EHR data used in this study. However, we have released the DDIWAS R package on GitHub (https://github.com/pwatrick/ddiwas) under an Apache License and archived version 0.1 on Zenodo.53 We have written a tutorial to extract data from EHR databases organized using the OHDSI/OMOP Common Data Model,52 which can be found at https://pwatrick.github.io/ddiwas/articles/extract_ehr_data.html. We have also prepared a guide to process and analyze extracted EHR data, which can be found at https://pwatrick.github.io/ddiwas/articles/ddiwas_r_package_tutorial.html.