Abstract
Ovomucin, a hen egg white protein, is characterized by its hydrogel-forming properties, high molecular weight, and extensive O -glycosylation with a high degree of sialylation. As a commonly used food ingredient, we explored whether ovomucin has an effect on the gut microbiota. O- Glycan analysis revealed that ovomucin contained core-1 and 2 structures with heavy modification by N -acetylneuraminic acid and/or sulfate groups. Of the two mucin-degrading gut microbes we tested, Akkermansia muciniphila grew in medium containing ovomucin as a sole carbon source during a 24 h culture period, whereas Bifidobacterium bifidum did not. Both gut microbes, however, degraded ovomucin O -glycans and released monosaccharides into the culture supernatants in a species-dependent manner, as revealed by semi-quantified mass spectrometric analysis and anion exchange chromatography analysis. Our data suggest that ovomucin potentially affects the gut microbiota through O -glycan decomposition by gut microbes and degradant sugar sharing within the community.
Keywords: Akkermansia muciniphila, Bifidobacterium bifidum, gut microbiota, mucin degradation, O-glycan, ovomucin
Abbreviations
CBB, Coomassie Brilliant Blue; CFU, colony forming unit; DMSO, dimethyl sulfoxide; Fuc, L-fucose; Gal, galactose, GalNAc, N -acetylgalactosamine; GlcNAc, N -acetylglucosamine; GlcNAc-6S, 6-sulfated N -acetylglucosamine; LNFP I, lacto- N -fucopentaose I; MALDI-TOF/MS, matrix assisted laser desorption ionization-time of flight/mass spectrometry; NeuAc, N -acetylneuraminic acid; NMR, nuclear magnetic resonance; PGM, porcine gastric mucin; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; S-Le a , sulfo-Lewis a trisaccharide.
INTRODUCTION
The hen egg white ovomucin, a mucin-like glycoprotein complex representing 3.5 % of hen egg white proteins, 1) is responsible for the gel property of albumen. With its viscosity, egg white provides mechanical protection from bacterial penetration into the egg yolk. 2) As a functional ingredient in food processing, ovomucin contributes to the foaming and emulsion properties of albumen. 3) Ovomucin is also known to have anti-viral, 4) 5) 6) 7) antiadhesive, 8) and anti-tumor activities. 9) Therefore, its application can be expected in a broad range of purposes, especially with the recent advances in separation techniques of ovomucin from other egg white components. 1) 7) 10) 11) 12)
Ovomucin is composed of two subunits: the relatively homogeneous, carbohydrate-poor α-ovomucin with a molecular size of 220-270 kDa, and the heterogeneous, carbohydrate-rich β-ovomucin with a molecular size of >400 kDa. 13) The carbohydrate contents for each subunit account for approximately 15 % and 58 % by weight, respectively. 14) They form a linear polymeric structure via disulfide bonds in a specific subunit ratio, often becoming insoluble as a result. α-Ovomucin, whose homologs are not found in mammals, has a 2,087 amino acid long polypeptide. 15) It has a similar protein structure to gel-forming mucins, with four von Willebrand D (VWD) domains and a cysteine-knot (CK) domain at the C-terminal end, which is involved in intermolecular oligomerization. However, α-ovomucin does not contain an obvious proline/threonine/serine-rich (PTS) domain, 16) which generally serves as the site of modification by O -glycans via GalNAc- O -Ser/Thr residues. It is modified by N -glycans at 15 potential N -glycosylation sites. 17) β-Ovomucin is an ortholog of a gel-forming mucin, MUC6, 16) and is abundantly modified by O -glycosylation, and also by N -glycosylation to a lesser degree. The 13 structures of ovomucin O -glycan have been suggested by NMR and mass spectrometric analyses. 18) 19) 20) 21) Those oligosaccharide moieties consist of 2–6 carbohydrate residues, which contain terminal capping by sialic acid and/or modification with a sulfate group.
In addition to antiviral and anti-adhesive activities, O -glycans of ovomucin can be a quality carbohydrate source for some bacteria, including specific gut microbes. Such mucin-degrading gut microbes in humans include Akkermansia muciniphila, which is notable for its association with reduction of obesity and type-2 diabetes, 22) 23) and Bifidobacterium bifidum , a probiotic species. Previous studies have shown that these species possess sets of extracellular glycosidases involved in mucin glycan degradation. 24) 25) 26) 27) Moreover, the sugars released by their glycosidases are not only assimilated by these species but are also consumed by specific microbes or shared within the bacterial community, 28) thus potentially influencing the structure of gut microbiota. Therefore, ovomucin can also be a potential agent to modify the gut microbiome through degradation by mucin-degrading gut microbes.
In this study, we analyzed ovomucin O -glycan structures by mass spectrometry and estimated the amount of each O -glycan species. Furthermore, to explore ovomucin’s capability as an agent to increase mucin-degrading gut microbes, A. muciniphila and B. bifidum were cultured in media containing ovomucin and examined for their ability to degrade ovomucin O -glycans. Our results indicated that mucin-degrading gut microbes can degrade ovomucin O -glycans for assimilation and release decomposed sugars into the environment, suggesting the possibility that ovomucin may have a significant effect on gut microbiota.
MATERIALS AND METHODS
Reagents.
Porcine gastric mucin (PGM, type III) was purchased from Sigma-Aldrich (St. Louis, MO, USA). PGM was extensively dialyzed against water and then lyophilized prior to use. Lacto- N -fucopentaose I (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc, LNFP I) and sulfo-Lewis a trisaccharide [(SO 3 − )-3Galβ1-3(Fucα1-4)GlcNAc, S-Le a ] were purchased from Carbosynth Ltd. (Compton, UK) and Prozyme, Inc. (Hayward, CA, USA), respectively. Unless otherwise stated, all other reagents used in this study were of analytical grade.
Preparation of enriched ovomucin fraction from egg white.
Lyophilized powder of the protein preparation containing ovomucin (hereafter referred to as ovomucin powder) was prepared and kindly provided by the R&D Division, Kewpie Corporation, Tokyo, Japan. Briefly summarized, the egg white was homogenized with a mixer, and diluted 2-fold with 100 mM NaCl, followed by 30 min of stirring. After adjusting the pH to 6.0 with 2 M hydrochloric acid, the protein precipitation was pelleted by centrifugation at 10,000 × G for 20 min. The protein pellet was redissolved in 100 mM NaCl, the pH was adjusted to 6.0, and the protein precipitation was pelleted as mentioned above, and this was repeated five times. The final protein precipitation was collected and lyophilized.
Glycoprotein analysis.
The ovomucin powder (100 μg) was dissolved in 45 μL of 43 mM sodium phosphate buffer (pH 7.5), and treated with 1 μL of peptide: N -glycanase F (PNGase F, Roche Diagnostics, Rotkreuz, Switzerland) and incubated for 18 h at 37 °C to remove N -glycans. The samples with and without PNGase F treatment were then separated on a 5–20 % acrylamide gel by SDS-polyacrylamide gel electrophoresis (PAGE). To detect the proteins, the gels were stained with Coomassie Brilliant Blue R-250 (Quick-CBB, Wako Pure Chemical, Osaka, Japan). For the detection of glycoproteins, the gels were stained using ProQ Emerald 300 Glycoprotein Gel and Blot Stain Kit (Thermo Fischer Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. Stained gels were visualized using a 312 nm UV transilluminator.
Release and purification of O-glycans from mucin.
To collect ovomucin glycoproteins from culture media after incubation with or without gut microbes (described later), ice-cold acetone (4-fold volume of the culture media) was added and placed on ice for 20 min. Then, the protein precipitation was collected by centrifugation at 15,000 × G for 20 min at 4 °C. O -Linked glycans were released by reductive β-elimination under the following conditions: the protein precipitation was resuspended in 1 mL of 1 M sodium borohydride and 100 mM sodium hydroxide and incubated for 18 h at 45 °C. The reaction was then stopped by adding 10 % acetic acid on ice, followed by the addition of glycan standards (500 pmol of LNFP I and 250 pmol of S-Le a per 100 μg mucin protein). Next, the solution was applied onto a column of Dowex 50WX8 (H + form, Sigma-Aldrich) resin (approximately 1 mL bed volume), washed out with 5 % acetic acid, and then lyophilized to dryness. Residual borate was removed as an azeotrope with methanol by adding 0.3 mL of 10 % acetic acid in methanol and drying under a nitrogen stream, which was repeated two additional times. To remove residual peptides and reagent contaminants, the dried material was resuspended in 500 μL of 5 % acetic acid and loaded onto a Sep-Pak C 18 cartridge column (100 mg, Waters, Milford, MA, USA) pre-equilibrated with at least 3 mL of 5 % acetic acid. Released oligosaccharide alditols were recovered by collecting the column run-through and an additional 2 mL of wash with 5 % acetic acid and lyophilized. For further clean up, the samples were dissolved in 1 mL of ultrapure water and load onto a graphitized carbon (GC) cartridge column (InertSep GC column, 150 mg/3 mL, GL Sciences, Tokyo, Japan). GC cartridge columns were activated by 12 mL of 0.1 % trifluoroacetic acid in 80 % acetonitrile and washed with 8 mL of ultrapure water before use. After loading the samples, columns were washed with 1 mL of ultrapure water. The O -glycan alditols were eluted in 25 % (v/v) acetonitrile containing 0.05 % (v/v) trifluoroacetic acid.
Glycan permethylation and phase-separation.
The purified oligosaccharide alditols were permethylated as described previously. 29) In addition to hydroxy groups, carboxy groups and secondary amines of N -acetyl groups were also methylated with this method. Briefly, the glycan samples were reconstituted in 200 μL of DMSO. Then, 250 μL of a base (a mixture of saturated sodium hydroxide and DMSO) and 100 μL of methyl iodide were added, and the mixtures were vigorously mixed for 5 min. After that, 2 mL of 5 % acetic acid were added and mixed by pipetting until the cloudiness became clear. Subsequently, dichloromethane (2 mL) was added, vortexed, and centrifuged (470 × G , 3 min at room temperature) and the upper layer (water-phase), which contained the permethylated sulfated glycan alditols, was transferred to another tube. This phase-separation was repeated four times. At last, the upper layer was removed and the lower dichloromethane layer (organic phase) containing the permethylated non-sulfated glycan alditols was dried under a nitrogen stream. For further purification, both glycan fractions were load on to a Sep-Pak C 18 cartridge column pre-equilibrated with water and eluted in methanol. Permethylated glycan alditols were dried under a nitrogen stream.
Mass spectrometric analysis of permethylated O-glycan alditols.
The permethylated glycans were dissolved in 2,5-dihydroxybenzoic acid matrix solution (10 mg/mL 2,5-dihydroxybenzoic acid in 50 % methanol) and subjected to MS analysis. MALDI-TOF/MS or MALDI-TOF/TOF/MS (LIFT) analyses were implemented by using AutoflexIII (Bruker Daltonics, Billerica, MA, USA) in the positive ion mode and the reflector mode. Glycan compositions were estimated based on the m/z values of glycan ion peaks by using the ExPASy GlycoMod tool (https://web.expasy.org/glycomod/). MS data was acquired in a m/z range from 400 to 3,000.
Bacterial strains and culture conditions.
Akkermansia muciniphila JCM 30893 and Bifidobacterium bifidum JCM 1254, which were obtained from RIKEN BioResource Research Center (Ibaraki, Japan), were pre-cultured at 37 °C in Gifu Anaerobic Medium (GAM) (Nissui Pharmaceutical, Tokyo, Japan) under anaerobic conditions using the AnaeroPack system (Mitsubishi Gas Chemical Co., Tokyo, Japan). Basal medium (2 g/L yeast extract, 10 g/L peptone, 5 g/L sodium acetate, 2 g/L diammonium citrate, 0.2 g/L magnesium sulfate, 2 g/L dipotassium hydrogen phosphate, and 1 g/L cysteine hydrochloride), which was supplemented with a reducing reagent (0.8 g/L cysteine hydrochloride and 4.4 g/L sodium carbonate at final concentrations) after autoclaving, was used for culture. 30) Ovomucin powder or porcine gastric mucin (PGM) was added to the medium at 0.5 % (w/v) as a carbon source. The growth was monitored by determining the number of colony-forming units (CFU) on GAM agar plates at the indicated time points. Data were expressed as means ± standard errors (SE).
Monosaccharide analysis.
High–performance anion exchange chromatography (HPAEC) with pulsed amperometric detection (PAD) was implemented using Dionex ICS-3000 (Thermo Fisher Scientific) with the CarboPac PA1 column (2 × 250 mm, Dionex) at 30 ºC. Eluting solutions used were ultrapure water (mobile phase A), 0.25 M NaOH (mobile phase B), and 1 M sodium acetate (mobile phase C). For separation of neutral monosaccharides including Fuc, Gal, GalNAc, and GlcNAc, the elution was performed at a flow rate of 0.25 mL/min with an isocratic eluent of 94.0 % A/5.5 % B/0.5 % C for the first 20 min. The column was then washed with an eluent of 19.5 % A/80 % B/0.5 % C for next 10 min and equilibrated with the initial eluent of 94.0 % A/5.5 % B/0.5 % C for 15 min before the next run. For detection of NeuAc, the elution was carried out as described previously. 31) The standard curves were created with the known concentrations of the sugars.
Acid hydrolysis to release sialic acid residues.
For quantification of the total amount of NeuAc in the protein preparation, NeuAc residues were released by acid hydrolysis. The sample was incubated in 0.01 M hydrochloric acid for 1 h at 80 °C and followed with neutralization by adding an equal volume of 0.01 M sodium hydroxide. The samples were subjected to monosaccharide analysis by HPAEC-PAD.
RESULTS
Assessment of ovomucin sample.
Hen egg white is known to contain ovalbumin (540 g/kg egg white protein), ovotransferrin (120 g/kg), ovomucoid (110 g/kg), ovomucin (35 g/kg), and lysozyme (35 g/kg), as major protein fractions as well as ovomacroglobulin (5 g/kg) and other proteins as minor protein fractions. 1) The ovomucin powder, which was prepared as described in the Materials and Methods section, was first separated on SDS-gradient polyacrylamide gel electrophoresis and stained with Coomassie Brilliant Blue ( Fig. 1A ). The two major bands with approximately 270 kDa and 150 kDa in size were presumed to be α-ovomucin and ovomacroglobulin, respectively. 1) 7) 32) The broadly stained larger band (over 400 kDa), which hardly migrated into the gel, was presumed to be β-ovomucin, some of which is present as an insoluble polymeric complex of α- and β-ovomucin. The minor smaller bands are considered to be ovalbumin (about 50 kDa) and lysozyme (14 kDa). Upon PNGase F treatment for removal of N -glycans, the band of ovomacroglobulin slightly shifted due to the loss of N -glycans, while the band of α-ovomucin was mostly unaffected. Supposing that α-ovomucin is modified with di-sialylated N -glycans at all 15 potential sites, 17) removal of these N -glycans would cause its molecular weight to decrease by 34 kDa. The high molecular weight of this protein may make it difficult to detect the band shift in SDS-PAGE analyses. It is also possible that the negative charge of sialylated glycans attached to the non-treated native protein affects migration in the gel during electrophoresis. When the gels were stained using periodate-based ProQ Emerald 300 Gel stain kit for the detection of glycoproteins, both α- and β-ovomucin molecules were stained regardless of treatment with PNGase F, whereas the staining of ovomacroglobulin became faint after PNGase F treatment ( Fig. 1B ). The periodate staining for other smaller proteins was negligible. Together, these results indicate that the O -glycans liberated from ovomucin powder in this study represent ovomucin glycoproteins.
Fig. 1. Glycoproteins in the protein preparation used in this study.
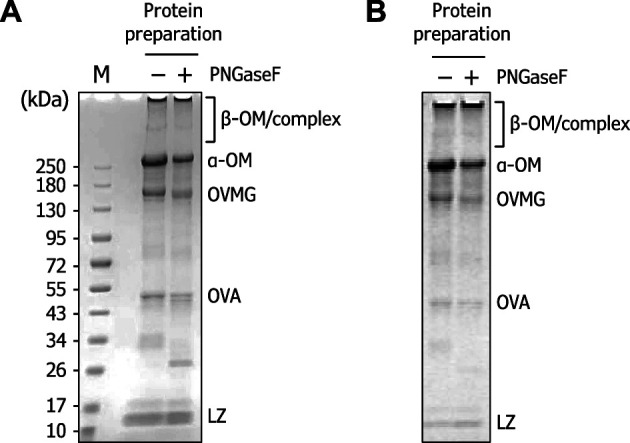
(A) CBB staining of the sample (10 μg) separated on a 5–20 % acrylamide gel. Prior to separation, PNGase F treatment was performed. (B) Periodate-based ProQ Emerald 300 staining of the sample. OM, ovomucin; OVMG, ovomacroglobulin; OVA, ovalbumin; LZ, lysozyme.
O -Glycan analysis of ovomucin.
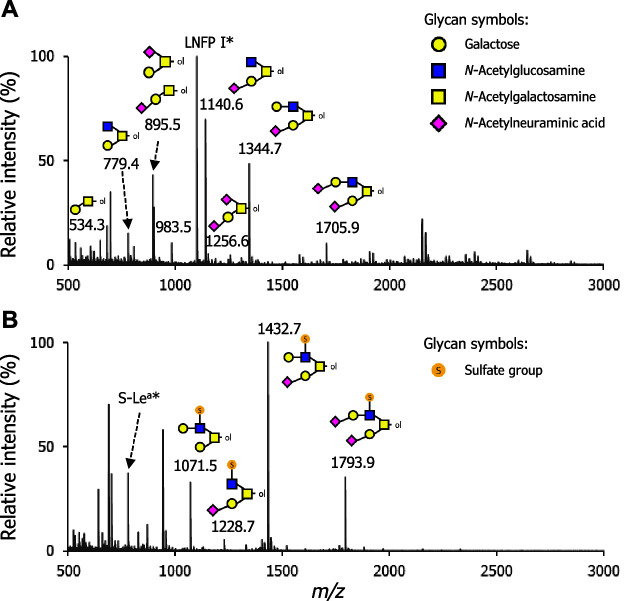
With the ovomucin powder, we analyzed the O -glycan profiles by mass spectrometry. The fractions of non-sulfated and sulfated permethylated O -glycan alditols, which are the result of glycan permethylation and phase-separation described in the Materials and Methods section, were separately subjected to MALDI-TOF/MS analysis in the positive ion mode. As shown in Fig. 2 , the glycan ion peaks corresponding to the compositions of permethylated glycan alditols as sodium adducts were identified in the full mass profiles: eight from the non-sulfated O -glycan fraction and four from the sulfated O -glycan fraction, summarized in Table 1 .
Fig. 2. Full mass profiles of the permethylated O -glycan alditols released from ovomucin.
O -Glycans were released through reductive β-elimination. The purified O -glycan alditols were permethylated and subsequently partitioned into an organic phase containing non-sulfated O -glycan alditols and a water-phase containing sulfated O -glycan alditols. Each fraction was further purified and subjected to analysis with MALDI-TOF/MS in a positive ion mode for (A) non-sulfated O -glycans and for (B) sulfated O -glycans. Deduced glycan structures are shown near the respective glycan ion peaks. Glycan symbols are depicted according to the nomenclature committee of the Consortium for Functional Glycomics (http://www.functionalglycomics.org/static/consortium/consortium.shtml). Asterisks indicate LNFP I ( m/z 1100.6 [M + Na] + ) and S-Le a ( m/z 780.3 [M + 2Na – H] + ), which were externally added to the specimens for the semi-quantification of glycan amounts.
Table 1.
O -Glycans from hen egg ovomucin before and after cultivation with mucin-degrading gut microbes.
| Deduced glycan composition a | Observed mass( m/z ) | Calculatedmonoisotopicmass( m/z ) | Core-type | Estimated amounts of O -glycans(pmol per 100 μg protein) b | ||
|---|---|---|---|---|---|---|
| Control | A. muciniphila | B. bifidum | ||||
| (Non-sulfated O -glycans) | [M + Na] + | |||||
| H1-HexNAcitol | 534.29 | 534.2885 | core-1 | 53.9 | 128.1 | 227.0 |
| H1N1-HexNAcitol | 779.41 | 779.4148 | core-2 | 76.1 | 53.7 | 52.6 |
| NA1H1-HexNAcitol | 895.46 | 895.4621 | core-1 | 215.7 | 48.3 | 76.8 |
| H2N1-HexNAcitol | 983.50 | 983.5146 | core-2 | 53.2 | 15.6 | 10.6 |
| NA1H1N1-HexNAcitol | 1140.58 | 1140.5885 | core-2 | 348.9 | 62.8 | 29.0 |
| NA2H1-HexNAcitol | 1256.62 | 1256.6358 | core-1 | 24.0 | 5.6 | 3.9 |
| NA1H2N1-HexNAcitol | 1344.69 | 1344.6882 | core-2 | 242.4 | 41.5 | 9.3 |
| NA2H2N1-HexNAcitol | 1705.86 | 1705.8619 | core-2 | 51.7 | 17.8 | 3.3 |
| (Sulfated O -glycans) | [M + 2Na − H] + | |||||
| S1H2N1-HexNAcitol | 1071.56 | 1071.4377 | core-2 | 220.1 | 26.6 | 25.2 |
| S1NA1H1N1-HexNAcitol | 1228.66 | 1228.5116 | core-2 | 36.5 | 28.3 | 7.6 |
| S1NA1H2N1-HexNAcitol | 1432.76 | 1432.6114 | core-2 | 666.2 | 118.0 | 13.1 |
| S1NA2H2N1-HexNAcitol | 1793.94 | 1793.7850 | core-2 | 237.0 | 58.8 | 0.8 |
a Abbreviations used are as follows; hexose, H; N -acetylhexosamine, N; N -acetylneuraminic acid, NA; sulfate group, S. b Estimated amounts of each O -glycan species were calculated based on the peak intensities normalized with those of externally added standards (LNFP I for the non-sulfated O -glycans and S-Le a for the sulfated O -glycans)(see Figs. 2 and 4 ).
MS/MS fragments for each glycan ion peak were further obtained by MALDI-TOF/TOF/MS (LIFT) analysis. The pertinent diagnostic ions determining each glycan structure, i.e. m/z 620 for a fragment of N -acetylneuraminic acid-substituted galactose (NeuAc-Gal) as a sodium adduct, were detected in the MS/MS profiles, and the deduced glycan structures are shown in Fig. S1 and S2 (see J. Appl. Glycosci. Web site).
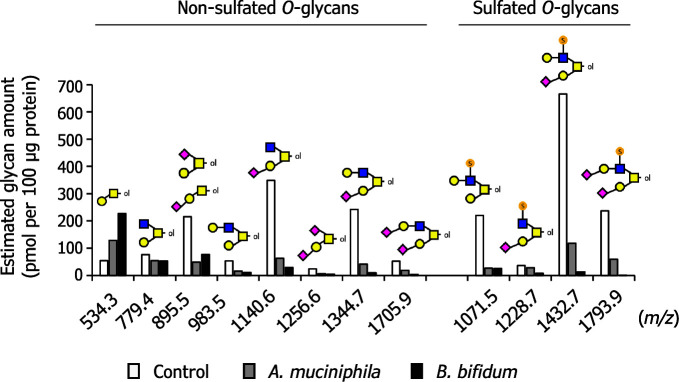
Furthermore, semi-quantification for each glycan species was performed based on the ion intensities by normalization with those of the externally added glycan standards: LNFP I for non-sulfated glycans and S-Le a trisaccharide for sulfated glycans (see Fig. 4 , control). The most abundant glycan was a monosialylated core-2 structure with a monosulfated LacNAc unit ( m/z 1,432.8 [M + 2Na − H] + , estimated amount: approximately 670 pmol per 100 μg protein), followed by a monosialylated core-2 structure ( m/z 1,140.6 [M + Na] + , estimated amount: approximately 350 pmol per 100 μg protein) and a monosialylated core-2 structure with a non-sulfated LacNAc unit ( m/z 1,344.7 [M + Na] + , estimated amount: approximately 240 pmol per 100 μg protein). Based on this semi-quantification data and the deduced glycan structures, the molar ratio of the compositional monosaccharides in total ovomucin O -glycans was calculated to be 4.8 (Gal), 1.0 (GlcNAc), 2.9 (GalNAc), 2.8 (NeuAc), and 1.5 (GlcNAc-6S), respectively.
Fig. 4. Ovomucin O -glycans were degraded by mucin-degrading gut microbes.
Akkermansia muciniphila JCM 30893 and Bifidobacterium bifidum JCM 1254 were cultured in media supplemented with 0.5 % ovomucin powder as a sole carbon source. As a control, the same medium was incubated without bacteria. After 24 h culture, O -glycans were analyzed with MALDI-TOF/MS (Fig. S1; see J. Appl. Glycosci. Web site). Semi-quantification of each glycan species was carried out with normalization by use of peak intensities of LNFP I (500 pmol added) and S-Le a (250 pmol added) for the non-sulfated O -glycans and the sulfated O -glycans, respectively.
In summary, ovomucin O -glycans were found to comprise two series of O -glycans based on core-type: core-1 structures and core-2 structures that are modified by mono- or di-sialylation with NeuAc. Core-1 series contain a sialyl T-antigen. The sulfate group is added onto GlcNAc residue on the LacNAc moiety of core-2 structures. We failed to detect a monosialylated core-1 structure with 6-sulfated GalNAc-moiety reported in a previous study by Kato et al. 19) Nonetheless, the present data were essentially consistent with the previous reports of O -glycan analysis of ovomucin, 18) 19) 20) 21) which demonstrates the validity of our results.
Akkermansia muciniphila growth on ovomucin.
In order to examine whether ovomucin O -glycan supports the growth of mucin-degrading gut microbes, we cultured Akkermansia muciniphila and Bifidobacterium bifidum in media containing 0.5 % (w/v) ovomucin powder as a sole carbon source and determined colony-forming units (CFU) after 24 h of cultivation. As a result, the CFU of A. muciniphila increased 6.9-fold ( p < 0.05, two-tailed t -test) whereas no apparent increase was observed for B. bifidum ( Fig. 3 ). Both bacteria grew well with 0.5 % PGM, which was used as an alternative carbon source and is known to be abundantly fucosylated. 33) Neither of species grew without glycoprotein supplementation.
Fig. 3. Growth of mucin-degrading gut microbes on ovomucin.
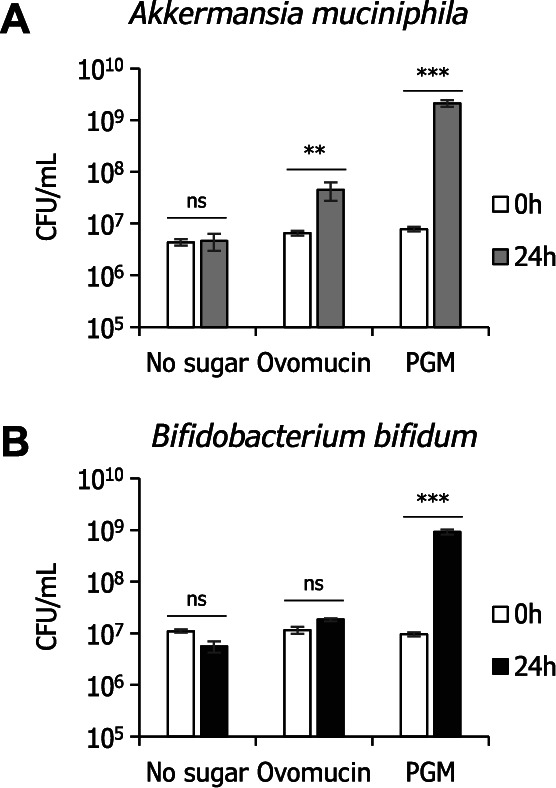
Akkermansia muciniphila JCM 30893 (A) and Bifidobacterium bifidum JCM 1254 (B) were cultured in basal media containing 0.5 % ovomucin powder or 0.5 % porcine gastric mucin (PGM) as a sole carbon source. Colony-forming units (CFU) were determined at 0 h and 24 h of incubation time. Mean values are shown with ± SE. *** P < 0.001; ** P < 0.01; ns, not significant ( n = 3, two-tailed t -test).
O -Glycan degradation by gut microbes.
The O -glycans after the 24 h of incubation were then examined and compared with respect to the estimated amounts for the identified twelve O -glycan species (Fig. S3; see J. Appl. Glycosci. Web site and Fig. 4 ). As a result, in both bacterial cultures, all the glycan peaks were markedly reduced after incubation except for those of unmodified core-1 structures (Galβ1-3GalNAc-itol, m/z 534.3), which is generated by the degradation of larger glycan structures. B. bifidum seemed to degrade a slightly higher amount of more complex O -glycans. These results indicated that overall rates of glycan decomposition were similar between the two bacteria.
Amounts of free monosaccharides in the spent media.
We next determined the amounts of free monosaccharides (Gal, GalNAc, GlcNAc, NeuAc, and Fuc) in the spent media of 24 h cultures by HPAEC-PAD. GlcNAc-6S was not included in the analysis because the compound did not give a peak under the tested elution conditions. As shown in Fig. 5A , Gal, GlcNAc, and NeuAc were detected in the ovomucin-supplemented culture supernatant of A. muciniphila at concentrations of 37, 24, and 189 μM, respectively, whereas the concentrations in cultures with B. bifidum were 76, 0, and 120 μM, respectively. GalNAc and Fuc were not detected in either culture supernatants. Considering that similar degrees of degradation of ovomucin O -glycans were detected in the MS analysis between the two bacteria, the difference in the concentration of free monosaccharides may reflect the metabolic capacity of those bacteria for the monosaccharides. The initial NeuAc amount attached to ovomucin sugar chains was estimated to be 13.7 nmol/100 μg protein by acid hydrolysis followed by HPAEC-PAD analysis. On the other hand, the amounts of free NeuAc present in the supernatants in A. muciniphila- and B. bifidum- cultures were 3.8 nmol and 2.4 nmol/100 μg ovomucin, respectively ( Fig. 5B ). In the supernatant of PGM-containing medium cultured with A. muciniphila , neither NeuAc nor Fuc were detected, while low amounts of Gal and GlcNAc were found. In contrast, Gal, Fuc, and NeuAc, but not GlcNAc, were abundantly present in the spent media of PGM with B. bifidum ( Fig. 5C ). Consistent results were obtained by thin-layer chromatography analysis of free sugars of culture supernatants (Fig. S4; see J. Appl. Glycosci. Web site).
Fig. 5. Amounts of free monosaccharides in the culture supernatants.
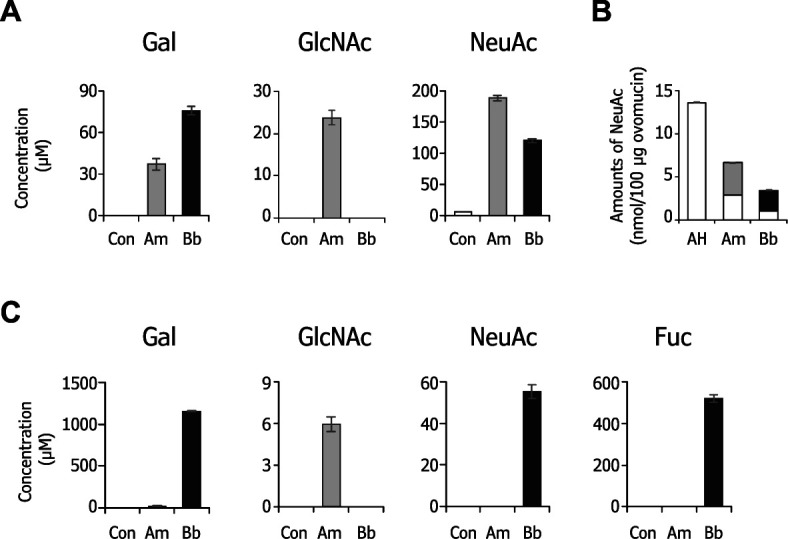
The monosaccharides in the spent media after 24 h cultivation with Akkermansia muciniphila (Am), Bifidobacterium bifidum (Bb), and control with no bacteria (Con) in the presence of 0.5 % (w/v) ovomucin powder and PGM were analyzed by HPAEC-PAD. Concentrations of free Gal, GlcNAc, and NeuAc in the ovomucin-containing media (A), and those plus Fuc in the PGM-containing media (C) are shown. Amounts of NeuAc released from ovomucin by incubation with Akkermansia muciniphila (gray) and Bifidobacterium bifidum (black) are compared with amounts released by acid hydrolysis (AH) treatment (B). Calculated amounts of residual NeuAc on ovomucin after 24 h culture are also shown in the graph (white in Am and Bb). Data represent the means of biological triplicates ± SE.
DISCUSSION
In this study, we analyzed O -glycan structures of ovomucin and estimated the amount of glycans by semi-quantification using full-mass profiles of MALDI-TOF/MS analysis. MS and MS/MS fragment analyses allowed us to identify nine non-sulfated structures (eight ion peaks) and four sulfated structures. Although MS analysis is incapable of determining sugar anomeric configurations and linkage positions in depth, our results of sugar compositions and glycan sequences is essentially consistent with previous studies, in which the glycan structures were analyzed by techniques involving NMR, 18) 20) 21) indicating the common modification patterns of ovomucin. It should be mentioned that our semi-quantification analysis revealed for the first time that the most prevalent O -glycan structure is a monosialylated, monosulfated core-2 structure.
Most of these O -glycan structures were decomposed by mucin-degrading gut microbes used in this study ( Fig. 4 ). A. muciniphila and B. bifidum commonly possess glycoside hydrolases (GHs) relevant to mucin degradation, 25) 26) 27) which include GH29/GH95 α-fucosidases, 25) 34) 35) GH33 sialidases, 36) 37) 38) GH2 β-galactosidases, 39) and GH20 β- N -acetylhexosaminidases. 39) 40) Thus, ovomucin O -glycans would be degraded by sequential enzymatic hydrolysis of the glycosidic linkages by those GHs. There are, however, some differences in the O -glycan degradation pathways between these bacteria. One such difference resides in the final step of degradation. In A. muciniphila , the Galβ1-3GalNAc disaccharide core-1 structure (galacto- N -biose, or T-antigen) linked to Ser/Thr residues of the polypeptide is hydrolyzed by GH35 β-galactosidase to release Gal, leaving the innermost GalNAc on polypeptides. 41) On the other hand, in B. bifidum , the Galβ1-3GalNAc disaccharide unit can be released by GH101 endo-α- N -acetylgalactosaminidase. 27) Unfortunately, we failed to detect the GalNAc signal ( m/z 330) in this study because MS data was acquired in a m/z range from 400 to 3,000. Detection of Galβ1-3GalNAc by HPAEC-PAD was also impossible under the conditions we employed, because the disaccharide is quite unstable in alkaline conditions. 42) In addition to the difference in T-antigen degradation processes between the two organisms, another difference may reside in sulfated glycan degradation process. We recently found that bbhII , a GH20 gene in B. bifidum , encodes a sulfoglycosidase that can release GlcNAc-6S from mucin. 43) A previous study 44) has demonstrated that A. muciniphila uses sulfatases for sulfated glycans, and it is possible that the organism encodes an unidentified sulfoglycosidase, as its genome contains eleven GH20 homologs.
Interestingly, a comparison of the concentrations of free sugars in the ovomucin-containing culture supernatants between A. muciniphila and B. bifidum revealed differences in their metabolic capacities. When the amounts of free Gal and GlcNAc were compared, Gal concentration of B. bifidum supernatants was double that of A. muciniphila . Conversely, GlcNAc was undetectable in B. bifidum supernatants, whereas a significant amount of free GlcNAc was left in A. muciniphila supernatants. These results may reflect their carbohydrate preferences, implying that B. bifidum utilizes GlcNAc prior to Gal and NeuAc. The results from PGM culture supernatants more clearly showed those preferences, where Akkermansia consumed all the sugars except GlcNAc, while B. bifidum left sugars other than GlcNAc unconsumed. These carbohydrate preferences and the ratio of monosaccharide components of ovomucin O -glycans may explain the reason why A. muciniphila but not B. bifidum grew on the ovomucin medium, although we only observed a 24 h snapshot ( Fig. 3 ). It should be mentioned that the total monosaccharide concentration derived from ovomucin O -glycans does not exceed 0.08 % (w/v), as estimated from semi-quantification MS analysis of ovomucin attached NeuAc quantification. The growth of A. muciniphila on ovomucin may rely on peptide backbone degradation.
Sialylation is a characteristic of ovomucin, where at least 13.7 nmol NeuAc was contained in 100 μg ovomucin powder ( Fig. 5B ). Free NeuAc found in the culture supernatants of A. muciniphila and B. bifidum was about 28 and 18 %, respectively, of NeuAc released by acid hydrolysis ( Fig. 5B ). In the human intestine, it is known that free NeuAc released from host glycoconjugates is utilized by a number of bacteria, including multiple species of Clostridia , Bacteroides , Bifidobacterium , certain subspecies/serovars/strains of Vibrio cholerae , Ruminococcus gnavus , and Akkermansia muciniphila that have the nan clusters, nanA/K/E or nanLET. 45) By analysis with KEGG pathway, it was predicted that A. muciniphila has a N- acetylneuraminate lyase (NanA) that converts NeuAc to N -acetylmannosamine (ManNAc), which could lead to peptideglycan biosynthesis via GlcNAc. 25) By contrast, B. bifidum does not have genes responsible for NeuAc assimilation. To our surprise, however, our results were contrary to past studies in that even though B. bifidum degraded a slightly higher amount of O -glycans into monosaccharides from ovomucin than A. muciniphila did, the concentration of free NeuAc left in supernatant of B. bifidum was lower than that of A. muciniphila ( Figs. 4 and 5A ). Although the reason for this is still unknown, it may suggest that B. bifidum converts NeuAc to unidentified compounds by an unknown pathway, because it was able to grow on NeuAc-containing medium as a sole carbon source with an OD 600 of up to 0.2. 26) NeuAc liberated from mucin O -glycan by B. bifidum can be shared with Bifidobacterium breve that possesses the nanA cluster. 46) Also, past studies have reported that co-culturing of A. muciniphila with non-mucin degrading butyrate-producing bacteria Anaerostipes caccae , Eubacterium hallii , and Faecalibacterium prausnitzii in media containing mucin as a sole carbon source resulted in syntrophic growth and production of butyrate. 47) Thus, it is possible that released sugars from ovomucin can enhance the growth of other non-mucin degrading gut microbes.
Ovomucin O -glycan is easily decomposed by mucin-degrading gut microbes, but its effect on gut microbiota formation or alteration would comprise of complicated processes such as direct feeding of certain gut microbes, indirect-feeding of other bacteria (cross-feeding), 28) and competition over released monosaccharides among the microbial members. Nonetheless, our results suggest the potential for the application of ovomucin as a food additive.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
Supplementary Material
Acknowledgments
We thank Drs. Takeshi Nakano and Kentaro Ifuku at Kyoto University for the use of a MALDI-TOF/MS instrument. We also thank Ms. Miriam N. Ojima at Kyoto University for English proofreading. Protein preparation used in this study is a gift from Kewpie Corporation (Tokyo, Japan).
References
- 1).Chang C., Lahti T., Tanaka T., and Nickerson M.T.: Egg proteins: fractionation, bioactive peptides and allergenicity. J. Sci. Food Agric., 98, 5547–5558 (2018). [DOI] [PubMed] [Google Scholar]
- 2).Kovacs-Nolan J., Phillips M., and Mine Y.: Advances in the value of eggs and egg components for human health. J. Agric. Food Chem., 53, 8421–8431 (2005). [DOI] [PubMed] [Google Scholar]
- 3).Shan Y., Ma M., Huang X., Guo Y., Jin G., and Jin Y.: Simple pH treatment as an effective tool to improve the functional properties of ovomucin. J. Food Sci., 77, 740–745 (2012). [DOI] [PubMed] [Google Scholar]
- 4).Tsuge Y., Shimoyamada M., Watanabe K., Amaoa M.S., and Watanabe K.: Differences in hemagglutination inhibition activity against bovine rotavirus and hen newcastle disease virus based on the subunits in hen egg white ovomucin. Biosci. Biotechnol. Biochem., 60, 1505–1506 (1996). [DOI] [PubMed] [Google Scholar]
- 5).Xu Q., Shan Y., Wang N., Liu Y., Zhang M., and Ma M.: Sialic acid involves in the interaction between ovomucin and hemagglutinin and influences the antiviral activity of ovomucin. Int. J. Biol. Macromol., 119, 533–539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Tsuge Y., Shimoyamada M., and Watanabe K.: Bindings of ovomucin to newcastle disease virus and anti-ovomucin antibodies and its heat stability based on binding abilities. J. Agric. Food Chem., 45, 4629–4634 (1997). [Google Scholar]
- 7).Wang Z., Tu A., Tang D., and Shan Y.: Effectively preparing soluble ovomucin with high antiviral activity from egg white. Int. J. Biol. Macromol., 118, 504–510 (2018). [DOI] [PubMed] [Google Scholar]
- 8).Kobayashi K., Hattori M., Hara-Kudo Y., Okubo T., Yamamoto S., Takita T., and Sugita-Konishi Y.: Glycopeptide derived from hen egg ovomucin has the ability to bind enterohemorrhagic Escherichia coli O157:H7. J. Agric. Food Chem., 52, 5740–5746 (2004). [DOI] [PubMed] [Google Scholar]
- 9).Watanabe K., Tsuge Y., Shimoyamada M., Ogama N., and Ebina T.: Antitumor effects of pronase-treated fragments, glycopeptides, from ovomucin in hen egg white in a double grafted tumor system. J. Agric. Food Chem., 46, 3033–3038 (1998). [Google Scholar]
- 10).Omana D.A. and Wu J.: A new method of separating ovomucin from egg white. J. Agric. Food Chem., 57, 3596–3603 (2009). [DOI] [PubMed] [Google Scholar]
- 11).Abeyrathne E.D.N., Lee H.Y., and Ahn D.U.: Sequential separation of lysozyme, ovomucin, ovotransferrin, and ovalbumin from egg white. Poult. Sci., 93, 1001–1009 (2014). [DOI] [PubMed] [Google Scholar]
- 12).Geng F., Huang Q., Wu X., Ren G., Shan Y., Jin G., and Ma M.: Co-purification of chicken egg white proteins using polyethylene glycol precipitation and anion-exchange chromatography. Sep. Purif. Technol., 96, 75–80 (2012). [Google Scholar]
- 13).Itoh T., Miyazaki J., Sugawara H., and Adachi S.: Studies on the characterization of ovomucin and chalaza of the hen’s egg. J. Food Sci., 52, 1518–1521 (1987). [Google Scholar]
- 14).Robinson D.S. and Monsey J.B.: Studies on the composition of egg-white ovomucin. Biochem. J., 121, 537–547 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Watanabe K., Shimoyamada M., Onizuka T., Akiyama H., Niwa M., Ido T., and Tsuge Y.: Amino acid sequence of alpha-subunit in hen egg white ovomucin deduced from cloned cDNA. DNA Seq., 15, 251–261 (2004). [DOI] [PubMed] [Google Scholar]
- 16).Lang T., Hansson G.C., and Samuelsson T.: An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics, 7, Article number : 197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Offengenden M., Fentabil M.A., and Wu J.: N -glycosylation of ovomucin from hen egg white. Glycoconj. J., 28, 113–123 (2011). [DOI] [PubMed] [Google Scholar]
- 18).Strecker G., Wieruszeski J.-M.M., Martel C., and Montreuil J.: Complete 1H- and 13 C-n.m.r. assignments for two sulphated oligosaccharide alditols of hen ovomucin. Carbohydr. Res., 185, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- 19).Kato A., Hirata S., and Kobayashi K.: Structure of the sulfated oligosaccharide chain of ovornucint. Agric. Biol. Chem., 42, 1025–1029 (1978). [Google Scholar]
- 20).Strecker G., Wieruszeski J.M., Martel C., and Montreuil J.: Determination of the structure of sulfated tetra- and pentasaccharides obtained by alkaline borohydride degradation of hen ovomucin. A fast atom bombardment-mass spectrometric and 1 H-NMR spectroscopic study. Glycoconj. J., 4, 329–337 (1987). [Google Scholar]
- 21).Strecker G., Wieruszeski J.M., Cuvillier O., Michalski J.C., and Montreuil J.: 1 H and 13 C-NMR assignments for sialylated oligosaccharide-alditols related to mucins. Study of thirteen components from hen ovomucin and swallow nest mucin. Biochimie, 74, 39–51 (1992). [DOI] [PubMed] [Google Scholar]
- 22).Depommier, C., Everard A., Druart C., Plovier H., Van Hul M., Vieira-Silva S., Falony G., Raes J., Maiter D., Delzenne N.M., de Barsy M., Loumaye A., Hermans M.P., Thissen J.P., de Vos W.M., and Cani P.D.: Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med., 25, 1096–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., Guiot Y., Derrien M., Muccioli G.G., Delzenne N.M., de Vos W.M., and Cani P.D.: Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci., 110, 9066–9071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Shin J., Noh J.R., Chang D.H., Kim Y.H., Kim M.H., Lee E.S., Cho S., Ku B.J., Rhee M.S., Kim B.C., Lee C.H., and Cho B.K.: Elucidation of Akkermansia muciniphila probiotic traits driven by mucin depletion. Front. Microbiol., 10, 1137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Ottman N., Davids M., Suarez-Diez M., Boeren S., Schaap P.J., Martins Dos Santos V.A.P., Smidt H., Belzer C., and de Vos W.M.: Genome-scale model and omics analysis of metabolic capacities of Akkermansia muciniphila reveal a preferential mucin-degrading lifestyle. Appl. Environ. Microbiol., 83, e01014–e01017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Turroni F., Bottacini F., Foroni E., Mulder I., Kim J.-H., Zomer A., Sanchez B., Bidossi A., Ferrarini A., Giubellini V., Delledonne M., Henrissat B., Coutinho P., Oggioni M., Fitzgerald G.F., Mills D., Margolles A., Kelly D., van Sinderen D., and Ventura M.: Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci., 107, 19514–19519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Kiyohara M., Nakatomi T., Kurihara S., Fushinobu S., Suzuki H., Tanaka T., Shoda S. -i., Kitaoka M., Katayama T., Yamamoto K., and Ashida H.: α- N -Acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J. Biol. Chem., 287, 693–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Gotoh A., Katoh T., Ling Y., Sakanaka M., Yamada C., Asakuma S., Urashima T., Tomabechi Y., Katayama-ikegami A., Kurihara S., Yamamoto K., Harata G., He F., Hirose J., Okuda S., and Katayama T.: Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep., 8, 13958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Kumagai T., Katoh T., Nix D.B., Tiemeyer M., and Aoki K.: In-gel β-elimination and aqueous-organic partition for improved O - and sulfoglycomics. Anal. Chem., 85, 8692–8699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., and Kitaoka M.: Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem., 286, 34583–34592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Sakurama H., Kiyohara M., Wada J., Honda Y., Yamaguchi M., Fukiya S., Yokota A., Ashida H., Kumagai H., Kitaoka M., Yamamoto K., and Katayama T.: Lacto- N -biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J. Biol. Chem., 288, 25194–25206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Geng F., Huang X., Majumder K., Zhu Z., Cai Z., and Ma M.: Mass spectrometry and two-dimensional electrophoresis to characterize the glycosylation of hen egg white ovomacroglobulin. J. Agric. Food Chem., 63, 8209–8215 (2015). [DOI] [PubMed] [Google Scholar]
- 33).Sugiyama Y., Gotoh A., Katoh T., Yoshida E., Honda Y., Kurihara S., Ashida H., Kumagai H., Yamamoto K., Kitaoka M., and Katayama T.: Introduction of H-antigen structures onto oligosaccharides and sugar chains of glycoproteins using highly efficient 1,2-α-L-fucosynthase. Glycobiology, 26, 1235–1247 (2016). [DOI] [PubMed] [Google Scholar]
- 34).Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., and Yamamoto K.: Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology, 19, 1010–1017 (2009). [DOI] [PubMed] [Google Scholar]
- 35).Katayama T., Sakuma A., Kimura T., Makimura Y., Hiratake J., Sakata K., Yamanoi T., Kumagai H., and Yamamoto K.: Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase Family 95). J. Bacteriol., 186, 4885–4893 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Kiyohara M., Tanigawa K., Chaiwangsri T., Katayama T., Ashida H., and Yamamoto K.: An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology, 21, 437–447 (2011). [DOI] [PubMed] [Google Scholar]
- 37).Ashida H., Tanigawa K., Kiyohara M., Katoh T., and Yamamoto K.: Bifunctional properties and characterization of a novel sialidase with esterase activity from Bifidobacterium bifidum. Biosci. Biotechnol. Biochem., 82, 2030–2039 (2018). [DOI] [PubMed] [Google Scholar]
- 38).Huang K., Wang M.M., Kulinich A., Yao H.L., Ma H.Y., Martínez J.E.R., Duan X.C., Chen H., Cai Z.P., Flitsch S.L., Liu L., and Voglmeir J.: Biochemical characterisation of the neuraminidase pool of the human gut symbiont Akkermansia muciniphila. Carbohydr. Res., 415, 60–65 (2015). [DOI] [PubMed] [Google Scholar]
- 39).Miwa M., Horimoto T., Kiyohara M., Katayama T., Kitaoka M., Ashida H., and Yamamoto K.: Cooperation of β-galactosidase and β- N -acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology, 20, 1402–1409 (2010). [DOI] [PubMed] [Google Scholar]
- 40).Wang M., Zhang X.Y., Guo R.R., Cai Z.P., Hu X.C., Chen H., Wei S., Voglmeir J., and Liu L.: Cloning, purification and biochemical characterization of two β- N -acetylhexosaminidases from the mucin-degrading gut bacterium Akkermansia muciniphila. Carbohydr. Res., 457, 1–7 (2018). [DOI] [PubMed] [Google Scholar]
- 41).Guo B.S., Zheng F., Crouch L., Cai Z.P., Wang M., Bolam D.N., Liu L., and Voglmeir J.: Cloning, purification and biochemical characterisation of a GH35 beta-1,3/beta-1,6-galactosidase from the mucin-degrading gut bacterium Akkermansia muciniphila. Glycoconj. J., 35, 255–263 (2018). [DOI] [PubMed] [Google Scholar]
- 42).Chiku K., Nishimoto M., and Kitaoka M.: Thermal decomposition of β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-D-hexopyranoses under neutral conditions. Carbohydr. Res., 345, 1901–1908 (2010). [DOI] [PubMed] [Google Scholar]
- 43).Katoh T., Maeshibu T., Kikkawa K., Gotoh A., Tomabechi Y., Nakamura M., Liao W.-H., Yamaguchi M., Ashida H., Yamamoto K., and Katayama T.: Identification and characterization of a sulfoglycosidase from Bifidobacterium bifidum implicated in mucin glycan utilization. Biosci. Biotechnol. Biochem., 81, 2018–2027 (2017). [DOI] [PubMed] [Google Scholar]
- 44).Derrien M., Vaughan E.E., Plugge C.M., and de Vos W.M.: Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol., 54, 1469–1476 (2004). [DOI] [PubMed] [Google Scholar]
- 45).Juge N., Tailford L., and Owen C.D.: Sialidases from gut bacteria: a mini-review. Biochem. Soc. Trans., 44, 166–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Nishiyama K., Nagai A., Uribayashi K., Yamamoto Y., Mukai T., and Okada N.: Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe, 52, 22–28 (2018). [DOI] [PubMed] [Google Scholar]
- 47).Belzer C., Chia L.W., Aalvink S., Chamlagain B., Piironen V., Knol J., and de Vos W.M.: Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. MBio, 8, e00770–e00777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.