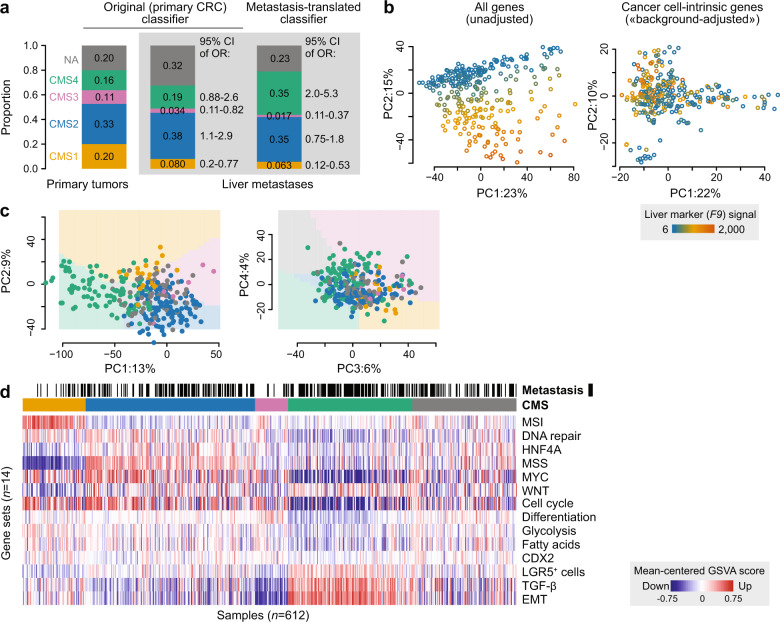
Fig. 2. Translation of CMS classification to liver metastases.
a Barplots illustrate differences in CMS distributions between primary tumors and liver metastases using the original primary CRC classifier and the new metastases-translated classifier. Odds ratios (ORs) indicate changes in proportions of each CMS compared to the primary setting (only counting confidently classified samples). b Principal component analyses demonstrate that the transcriptomic “liver background” (here illustrated by expression of the hepatocyte-enriched F9 gene marker) represents a large source of variation in raw expression data from bulk tissue samples of liver metastases (left), but not when only considering the 1104 cancer cell-intrinsic candidate markers after “liver background-adjustment” (right). c Principal component analyses showing CMS-classified liver metastases (colored dots, n = 295) projected onto the first four principal components from primary tumors. Colored backgrounds represent the dominant subtype in the primary tumors. d Gene set analysis with sample type (black: metastases; white: primary tumors) and assigned CMS (as in a) indicated on top. Samples are ordered according to CMS and p-values from the tailored CMS classifier. Gene sets (in rows) are ordered according to CMS hallmarks (CMS1 characteristics first, followed by CMS2 and so on).