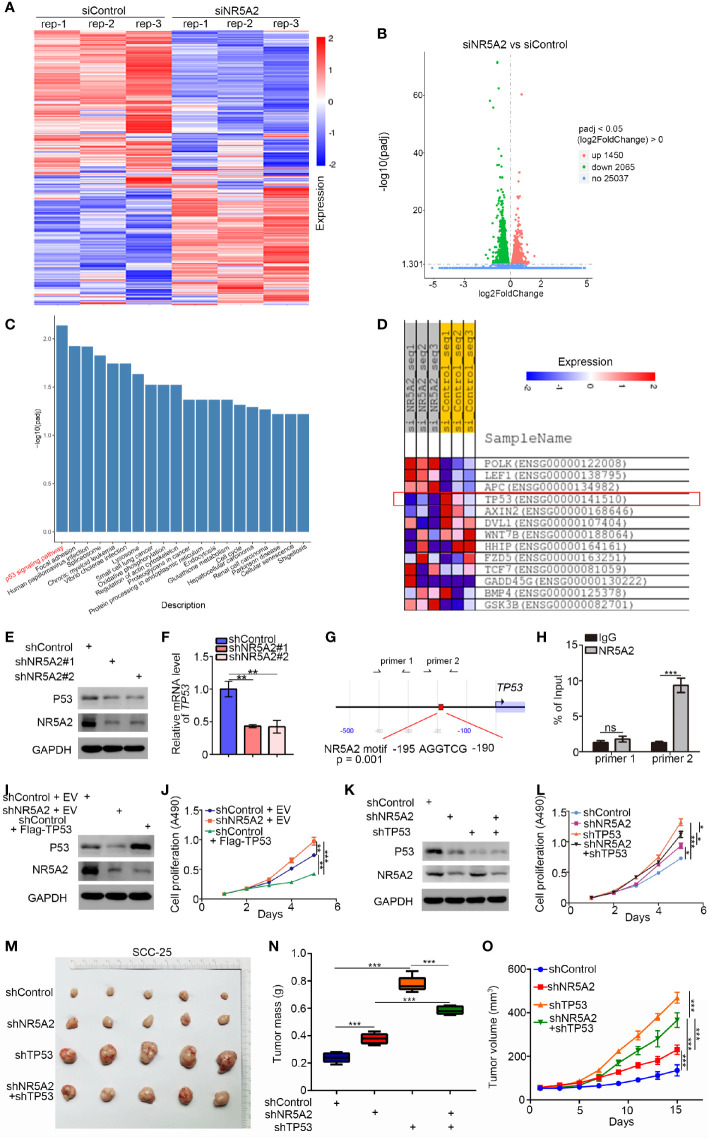
Figure 6.
NR5A2 regulates HNSCC cell growth in a p53-dependent manner. (A–D) heatmap (A) and volcano plot (B) to show the differential expressed genes of FaDu cells infected by siControl or siNR5A2. (C) Enrichment analysis suggests that the differential genes between siControl and siNR5A2 groups may related to HNSCC. (D) RNA-Seq data suggest that knocking down NR5A2 correlation with the decrease of p53 expression. (E, F), 48 h post-infection, FaDu cells were infected with shControl or shNR5A2 were harvested for Western blotting analysis (E) and RT-PCR analysis (F). The data shown are the mean values ± SD from three replicates. **P < 0.01. (G), the promoter region of TP53 was searched from The Eukaryotic Promoter Database exhibit that there was a consensus NR5A2 binding sites in the promoter region. (H), the ChIP-qPCR analysis of FaDu cells. The data shown are the mean values ± SD from three replicates. ***P < 0.001. (I, J), FaDu cells were infected with indicated shRNAs. After 48 h, cells were transfected with indicated plasmids (Empty vector (EV) and Flag-TP53) for another 24 h. Cells were harvested for Western Blot (I) and cell proliferation assay (J). The data shown are the mean values ± SD from three replicates. **P < 0.01; ***P < 0.001. (K–O), FaDu cells were infected with indicated shRNAs. 72 h post infection, cells were harvested for Western blotting (K), MTS assay (L), or xenografts assay (M). The tumor volume was measured every three days (N). Then the tumors were harvested, photographed (M) and weighed (O). For MTS assay, data are shown as means ± SD (n = 3). *P < 0.05, ***P < 0.001. For xenograft assay, data are shown as means ± SD (n = 5). ***P < 0.001.