Abstract

The development of a nanoparticle-based optical platform has been presented as a biosensor for detecting target-specific plant virus DNA. The binding dynamics of gold nanoparticles has been studied on the amine-functionalized surface by the attenuated total reflection (ATR)-based evanescent wave absorption method monitoring the localized surface plasmon resonance (LSPR). The developed surface was established as a refractive index sensor by monitoring the LSPR absorption peak of gold nanoparticles. This nanoparticle-immobilized surface was explored to establish as a biosensing platform with target-specific immunoglobulin (IgG) antibody–antigen interaction. The IgG concentration-dependent variation of absorbance was correlated with the refractive index change. After successfully establishing this ATR configuration as an LSPR-based biosensor, the single-stranded DNA of the chilli leaf curl virus was detected using its complementary DNA sequence as a receptor. The limit of detection of this sensor was determined to be 1.0 μg/mL for this target viral DNA. This ATR absorption technique has enormous potential as an LSPR based nano-biosensor for the detection of other begomoviruses.
Introduction
In recent years, the detection of harmful biomolecules or toxic elements/agents has been a major global challenge for public health and environmental and agricultural safety. The technological progress of portable integrated biosensors plays a critical role in the on-site detection of harmful biological substances. This biosensor helps to improve the severity through proper preventive tactics of disease management. In view of user convenience, several detection methodologies have been developed using various sensing platforms such as photonics,1,2 immunoassay,3,4 fluorescence-based colorimetry, and electrochemical methods.5,6 Among them, the photonic-based light–matter interaction technique is unique7−10 and immune to electromagnetic interference, which has the potential to revolutionize the biosensor as point-of-care device applications. Today’s most available biosensors work by measuring the change in light signals at a particular wavelength or over the band of spectra due to variation in the refractive index (RI) of the medium.11,12 The change in input light signal parameters such as transmitted/absorbed/reflected intensity, angular shift, wavelength shift, and phase-shift measurement is a tool to develop photonic-based compact and portable biomolecular sensors.12−15
Fiber optic sensors (FOSs)16−20 and attenuated total reflection (ATR) sensors21−24 have been proved to have enormous potential for the real-time detection of biomolecules utilizing the concept of the evanescent wave absorption method. Over the last few decades, the functionalized bare FOSs have been widely used to detect hazardous biomolecules with good sensitivity and selectivity. Similarly, the highly explosive chemicals and volatile gaseous molecules have been identified by ATR-infrared spectroscopy.25,26 Furthermore, the receptor-based functional molecule detection technique has also been explored in various active research areas to detect streptavidin,27,28 immunoglobulin (IgG),28−33 DNA,32,33 and biomarkers.34,35 However, for many biomolecules and micro-organisms, the available bare FOS was not able to detect the change in molecular absorption directly. Because, most of these molecules are composed of hydrocarbons, nitrogen, organo-sulfur, and oxygenated phosphorus compounds, which have optical absorption in the UV (wavelength < 300 nm) range. Therefore, the development of a cost-effective and hand-held instrument utilizing costly UV optical sources to monitor any biomolecular ligand-binding reaction using an FOS in the UV region remains a technological challenge. In another scenario, the ATR technique is very unique in the identification of characteristic vibrational spectra, but it also has limitations for portable device development such as achieving precious control of sample operating temperature, integration of infrared optics, and efficient detection with a miniaturized system.
To address the above problem, the FOS and ATR platforms have been modified to a surface plasmon resonance (SPR) system with various structures of metallic thin films. In this regard, novel metallic23,36−38 (Au, Ag, Al, Ti) thin films have been used as a plasmon-carrying layer on different substrates (e.g., glass, quartz, and so forth) to improve the sensor response. This SPR technique has been used to detect contaminated liquids, target specific binding of macromolecules and micro-organisms,39−43 where label-free detection, sensitivity, and limit of detection (LOD) have also been emphasized for sample quantification. However, the thin-film deposition system being an expensive process is the bottleneck for developing low-cost devices. Localized SPR (LSPR) has thus been introduced, which needs an easy chemical process for nanoparticle synthesis. This LSPR technique also enhances the sensitivity and has now become the forefront technique for better sample quantification in many optical sensors. According to Lorenz–Mie scattering theory, the light absorption efficiency of a spherical nanoparticle is strongly affected by the dielectric constant of the surrounding medium44,45 and its particle size.46−49 For uniform size distribution of nanoparticles, the absorption efficiency will increase with an increase in the RI of the surrounding medium, which will change the LSPR peak position as well as the absorption amplitude. This variation in LSPR signal is used to calculate the change in medium RIs and thus the sensor response is measured. To develop RI sensors, novel metal nanoparticles have been widely used in many optoelectronic biosensors,50−53 which are capable of producing plasmon resonance in the visible light spectrum (400 to 700 nm wavelength). Among all, it has been found that gold nanoparticles (AuNPs) are chemically more stable54,55 and offer a compatible multifunctional surface for the selective binding of a wide range of biological ligands.56−58 Recently, the use of AuNPs as an LSPR agent in both FOS and ATR platforms has been explored in many innovative ways for the detection of hazardous molecules, allergic proteins, carcinogenic nucleic acids, malignant cells, and viruses.20,59−62
Although the FOS platform provides better RI sensitivity (in the order of 10–5 to 10–6) compared to ATR (in the order of 10–4 to 10–5), FOS has certain constraints in the experimental process. FOS needs a large amount (100–200 μL) of samples, whereas ATR needs a very small amount (20–40 μL) samples. Since in many cases, the availability of biomolecular samples is less in quantity, our work is focused on the development of viral DNA detection protocols using the ATR-LSPR platform. In this work, the begomovirus DNA from the infected plants has been considered as the target biomolecule. The plant pathogens or plant viruses are one of the most important constraints in agriculture all over the world. Proper diagnosis is an essential aspect for the management of plant viruses.63−67 The polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) methods are popularly used techniques to detect plant viruses.68−72 However, these approaches are laboratory-dependent and time-consuming. Therefore, there is a need for on-site and rapid diagnostic techniques for the detection of plant viruses.
In the present work, the possibility of developing an evanescent wave absorption-based ATR-LSPR biosensor for the detection of plant viruses has been investigated utilizing the chilli leaf curl virus (ChiLCV; Genus: Begomovirus, Family: Geminiviridae) as a case representative of plant viruses. The amine-functionalized surface was used to immobilize the AuNPs by monitoring the nanoparticles’ LSPR absorbance peak. This surface was used to study the effect on the LSPR signal due to the change of RI using sucrose solutions and thus the sensing platform was calibrated for the RI sensor. Furthermore, the biomolecular binding was tested using a fluorescein isothiocyanate (FITC)-tagged target-specific IgG antigen. This antigen detection was confirmed by monitoring the LSPR absorbance peak at 540 nm wavelength. Also, the LOD of this IgG biomolecule was determined by varying the concentration of the target. Finally, this ATR-LSPR platform was transformed into a complementary oligonucleotide probe surface and the plant viral DNA, that is, the single-stranded DNA (ssDNA) of ChiLCV has been successfully detected.
Experimental Method
Optical Setup
An experimental setup for the absorbance measurement with ATR-LSPR configurations was developed using the Kretschmann prism configuration,22 as shown in Figure 1a. To facilitate biomolecular interactions, a circular (5 mm diameter) reaction chamber/cavity with the maximum available volume (40 μL) was prepared using a cellulose tape (Wonder-555). This sample cavity was attached on top of the functionalized coverslip and placed on the prism base surface using an RI matching (n = 1.515/20D) liquid. The reflected light from the prism base was focused using an objective lens into one end of the fiber port of an optical fiber coupler and another end of the fiber was connected to a spectrophotometer (AvaSpec-ULS2048CL-EVO). All experimental events were monitored in real time in the spectral range from 400 to 900 nm wavelength throughout the experiment. The detailed description of the experimental setup is given in Supporting Information S1.
Figure 1.
(a) ATR-LSPR experimental setup with a broadband light source, a prism, an objective lens and a spectrometer and (b) real-time-monitored LSPR absorbance peak evolution and AuNP binding time dynamics (inset figure).
LSPR Probe Preparation
The standard glass coverslip was functionalized by the silanization method,73 using 3-aminopropyl triethoxysilane (APTES) solution (Supporting Information S2, Figure S1) and placed on the prism surface to prepare LSPR probes. First, the absorption spectrum of de-ionized (DI) water was taken as a reference for AuNPs, later this DI was replaced by 40 μL AuNPs (∼50 nm size) solution (AuNP synthesis details are given in Supporting Information S2, Figure S2). The purple-red color of nanoparticles inside the sample cavity starts diminishing when nanoparticles began slowly attaching to the active amine sites (Supporting Information S2, Figure S3). It was observed that the LSPR absorption peak became sharp and prominent with time and no peak broadening or any secondary peak was observed throughout the immobilization period (Figure 1b). Initially, the absorbance dynamics of nanoparticles increased very fast and slowed down when it reached the steady state (inset image in Figure 1b). Then, the unbound AuNPs were removed carefully by washing the cavity with DI water without disturbing the LSPR peak and finally the final absorbance was found to be 0.42 units in DI water medium. The nanoparticle density was estimated from scanning electron microscopy (SEM; JEOL; JSM-7600F FEG-SEM) micrograph (Supporting Information S2, Figure S4) to be 117 to 130 nanoparticles/μm2.
Antibody (IgG) Immobilization
After immobilizing the AuNPs, 20 mM cysteamine (Cys) solution was introduced into the sample chamber for 10–15 min. The residual Cys was removed by washing with DI water and then the cavity was incubated with 5% glutaraldehyde (GA) solution for 15 min which led to a cross-linking reaction; however, one end of the GA chain was left open for human IgG (HIgG) receptors. First, the HIgG antibody (0.2 mg/mL) was immobilized for 30 min. After incubation, the excess amount of sample was removed and the cavity was washed with the phosphate-buffered saline (PBS) to remove any loosely bound molecules. The surface was then treated with bovine serum albumin (BSA) (4 mg/mL) to prevent any non-specific binding and again cleaned with PBS solution (details of the process are given in Supporting Information S3, Figure S5). The binding spectra were recorded and used for further analysis.
Complementary DNA Preparation
A small stretch of nucleotide sequence from the genome of ChiLCV was utilized as the receptor sequence to capture the target virus DNA. The receptor sequence contained 18 nucleotides (code: 862BM R, sequence: 5′ CATCAGAGCATTCTCACT 3′) which were in the complementary orientation of the corresponding sequence in the genome coordinate 976–993 of the virus (ChiLCV). The receptor sequence was synthesized from GCG Biotech, India. The length of the primer was found to be 70–74 base pairs with a length around 25 nm. Later, this receptor sequence was used for immobilization on the LSPR probe surface and thus complementary DNA (cDNA) probes were prepared.
Viral DNA (Target DNA) Isolation
Field samples with typical leaf curl symptoms were collected and tested by the PCR method with the specific primers of ChiLCV. From one of the positive viral samples, the virus was transmitted to healthy chilli seedlings through whitefly inoculation under controlled environmental conditions, and the stock culture was maintained in the greenhouse (Supporting Information S3, Figure S6a). The inoculated plants exhibited typical leaf curl symptoms by 2–3 weeks post-inoculation, and the infected chilli leaf samples (100 mg) showing typical leaf curl symptoms were used to isolate total DNA using the DNeasy Plant Mini Kit (Quiagen, Valencia, CA, USA), which was then eluted with 50 μL of autoclaved double distilled water following the procedure described by Senanayake et al.(74) The presence of viral DNA was verified by PCR and the final concentration of viral DNA was found to be 0.01 mg/mL.
Non-Target DNA Preparation
To test the specificity and cross-validation of the sensor, an unknown target DNA sequence was obtained from a healthy chilli plant leaf that was not infected by the virus. Chilli seedlings were raised in an environment-controlled plant growth chamber (Supporting Information S3, Figure S5b), and DNA was isolated from 100 mg of leaf tissues as described above. The final healthy DNA concentration was found to be 0.01 mg/mL.
Results and Discussion
RI Sensitivity
The RI sensitivity of the prepared LSPR
probe was investigated, as given in Figure S7 (Supporting Information S4), using solutions with RI values
in the range of 1.3320–1.3999 RIU (refractive index units).
To understand the RI response, 40 μL of eight different sucrose
solutions (w/v; 5, 10, 15, 20, 25, 30, 35, and 40%) were carefully
dropped inside the sample cavity sequentially and the real-time change
in LSPR spectra was recorded. The sample cavity was washed with DI
water after each experiment. The change in the LSPR absorbance spectrum
was observed (Figure 2a) due to the exposure to different RI solutions on the surface.
The upward shift in absorbance spectra was observed with respect to
the baseline (DI water as the reference) because of the medium RI
change46,59,75 around the
AuNPs. Furthermore, this shift in the spectrum was considered for
baseline correction by calculating the change in LSPR absorbance ΔA [ΔA = A540 – A800, where A540 and A800 are absorbances
at 540 and 800 nm wavelengths, respectively] and plotted (inset in Figure 2a) as a function
of RI n of sucrose solutions. As seen from the figure,
the change in absorbance follows a linear relation with the RI. The
stability of absorbance A540 was analyzed
as a function of time (t vs A540) by recording
the absorbance response for 180–200 s from one value to other
higher values of the sucrose solution (Figure 2b). The instant change in absorbance was
observed immediately after introducing the sucrose solution into the
chamber (inset image in Figure 2b). This is because the RI of the sucrose solution is different
from that of DI, which changes the surrounding medium RI near the
nanoparticles.20,21 The response time of LSPR was
found for the change of RI of the solution to be around 2–3
s. During this experiment, it was also observed that the absorbance
increased from 0.42 to 0.93 units (from the reference DI water, RI
= 1.3320, to the highest concentration of sucrose, RI = 1.3999). The
effect of RI on absorbance A540 of nanoparticles
due to exposure in RI solutions was calculated under a stable condition
(t = 180 s), and the percentile value of normalized
absorbance ΔA540Norm [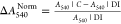 , where A540|C is the absorbance of the sucrose solution at
different concentrations and A540|DI is
the LSPR absorbance of immobilized AuNPs in the DI medium] is presented
in Figure 2c as a function
of the RI n of sucrose solutions. During each set
of experiments, the absorbance value A540|DI was maintained around 0.42 units to ensure identical immobilization
that maintained the density of AuNPs on the surface. As observed in
the figure, the ΔA540 is varied linearly from 20 to 105%
with the RI range n = 1.3418 to 1.3999 RIU. From
this study, the normalized absorbance ΔA540Norm was established
as a calibration eq 1 in our experimental results wherever absorbance-related RI was calculated
(used in biosensing). The slope or RI sensitivity (ΔA540/n) of this LSPR probe was evaluated and was found
to be 1514.45 ΔA540/RIU.
, where A540|C is the absorbance of the sucrose solution at
different concentrations and A540|DI is
the LSPR absorbance of immobilized AuNPs in the DI medium] is presented
in Figure 2c as a function
of the RI n of sucrose solutions. During each set
of experiments, the absorbance value A540|DI was maintained around 0.42 units to ensure identical immobilization
that maintained the density of AuNPs on the surface. As observed in
the figure, the ΔA540 is varied linearly from 20 to 105%
with the RI range n = 1.3418 to 1.3999 RIU. From
this study, the normalized absorbance ΔA540Norm was established
as a calibration eq 1 in our experimental results wherever absorbance-related RI was calculated
(used in biosensing). The slope or RI sensitivity (ΔA540/n) of this LSPR probe was evaluated and was found
to be 1514.45 ΔA540/RIU.
| 1 |
Figure 2.
(a) Obtained LSPR absorbance spectra of different sucrose solutions and the inset image represents the RI vs change in absorbance (n vs ΔA) for baseline correction, (b) absorbance response of different concentrations of sucrose solution with time (t vs A540) on the LSPR probe surface (monitored at 540 nm), and the extended plot shown in the inset image represents the abruptness in absorbance within t = 200–300 s, and (c) calculated normalized LSPR absorbance ΔA540Normvs RI of sucrose solutions (n vs ΔA540) for all samples after stable absorbance observed at t = 180 s.
The RI resolution R [R = Δn × ΔAmin/ΔA540Norm] of this ATR-LSPR configuration was found to be 7.04 × 10–6 RIU for the relative change in absorbance ΔA540 = 0.1348 within the RI range of 1.3478 to 1.3573 RIU (assuming the measurable absorbance ΔAmin = 0.0001 units). Recently, RI resolution has been demonstrated in the range from 1 × 10–6 to 3 × 10–8 RIU for the excitation of long-range surface plasmons of AuNPs by conventional intensity-modulation using the ATR method.7,60,76,77
Biosensor Development
The prepared AuNP probe surface was further used to detect the HIgG- and goat-anti-HIgG (GaHIgG)-based antigen–antibody biomolecular interaction by observing the LSPR peak kinetics. During this interaction, the LSPR absorption A540 was monitored, and the corresponding important parts of absorbance are labeled as illustrated in the sensogram (Figure 3a). As seen in the figure, in all experimental processes, wherever the terminology DI or DI + PBS is indicated, it means that they were used only for the purpose of obtaining the steady-state baseline before the incorporation of any analyte or as a cleaning buffer after the incubation. In process I (t = 0–3000 s), first, the nanoparticle-immobilized LSPR surface was prepared by incorporating a colloidal AuNP solution in the sample chamber. The immobilization was confirmed by observing the binding dynamics of AuNPs (the process is presented in the Experimental Section, Figure 1b). Then, in processes II (t = 3000–3826 s) and III (t = 3826–4864 s), the nanoparticle-immobilized surface was further functionalized using cross-linkers. First, they were functionalized by incubating with Cys dihydrochloride (40 μL of 20 mM concentration) and then with bi-functional GA (40 μL of 5% GA in DI water). The primary linker Cys acts as a cross-linking molecule where one end of the amine (−NH2) functional group is attached with the AuNP surface and the other end with an aldehyde (−CHO) functional group of GA. The thiol ligands of Cys can be replaced by the citrate groups from the surface of anionic AuNPs by electrostatic interaction.79 The other end of GA is left free for receptor molecules that guarantee that the prepared LSPR surface is ready for the HIgG receptor. In process IV (t = 4864–7610 s), the PBS analyte (10 mM, pH = 7.4) containing the HIgG (40 μL, 0.2 mg/mL concentration) antibody sample is introduced to prepare the receptor-immobilized surface. The real-time increase in absorbance due to HIgG antibody binding was observed compared to the buffer. This monoclonal HIgG binds to the cross-linked surface selectively via covalent immobilization78−82 and reached the LSPR absorbance plateau after 26 min. In the next process, process V (t = 7610–7810 s), the unreacted aldehyde surface or free binding sites were treated with BSA protein (40 μL, 4 mg/mL concentration) to prevent any non-specific binding and then cleaned with PBS buffer after 15 min. Now, this sensing surface is ready for the detection of target-specific molecules. Finally, in process VI (t = 7810–9230 s), the FITC-tagged Fc fragment-specific GaHIgG antigen solution (40 μL, 30 μg/mL concentration) was introduced into the sample chamber. It was observed that the absorbance signal increases exponentially due to specific binding with HIgG where a similar absorbance plateau was reached after 20 min and then unbound molecules were removed by rinsing with DI water.
Figure 3.
(a) Complete time-resolved absorbance response steps of HIgG–GaHIgG on the LSPR probe surface, (b) extended view of real-time absorbance dynamics of GaHIgG after incubation with 30 μg/mL concentration (starting point t = 0 s = t8160 s and t = 600 s = t8760 seconds), where ΔAb is the change in absorbance due to binding, and nt is the RI where absorbance ΔAb was calculated after 10 min of incubation, (c) time-resolved absorbance response of five different concentration (0.5, 1, 10, 30, and 50 μg/mL) of GaHIgG on LSPR probes monitored at 540 nm, (d) fluorescent microscope image of substates when different concentrations of GaHIgG samples were incubated at a fixed concentration of HIgG (0.2 mg/mL), (e) double Y axes graph of GaHIgG concentration vs observed absorbance at t – 10 min (con. vs At along the left axis; XY1) and GaHIgG concentration vs change in absorbance ΔAb was calculated (t = 10 min) after binding (con. vs ΔAb along right axis; XY2), (f) calculated change in RI Δnb surrounding the medium of immobilized AuNPs due to binding of GaHIgG (con. vs Δnb). Note: instant downward changes in absorbance are due to the introduction of analytes in the chamber.
Only for better illustration, the time-resolved absorbance response of the complete detection process step of GaHIgG is presented in Figure S8 (Supporting Information S4). After each incubation process, the red shift in the LSPR peak was observed, which confirmed the binding of cross-linkers, receptors, and target molecules (Supporting Information S4, Figure S9).
Two different time scales for the change in absorbance was observed in HIgG binding. Instant change in absorbance was observed immediately (within 3 s) after the HIgG sample was introduced inside the chamber. This change in absorbance was due to the different RI of the HIgG solution compared to the buffer (DI + PBS) solution. The absorbance dynamics of HIgG follow standard biomolecular association,83−87 where exponential growth in absorbance was observed.
In the case of GaHIgG, as observed in Figure 3b, the absorbance A540 was increased exponentially with time due to molecular affinity between the antigen and the antibody that follows the lock and key model.88,89 A similar kind of biomolecular binding was also observed in our previous work.19,20 This affinity between the receptor and the target is specific for the performance of the sensor. Because of the instant change in absorbance due to the RI of the GaHIgG solutions within 3 s, the actual binding kinetics was considered after 3 s of incubation, and the change in absorbance only due to binding (ΔAb) of GaHIgG was calculated using eq 2a.
| 2a |
where At is the peak absorbance at 540 nm wavelength, which was recorded with time t (the incubation of GaHIgG was considered for 10 min) and Abaseline is the baseline of DI + PBS buffer (after 3 s).
ΔAb depends on the concentration of GaHIgG and the incubation time. Therefore, the effect of concentration on LSPR absorption was found by varying GaHIgG concentrations from 50 to 0.5 μg/mL (Figure 3c). The concentration-dependent binding followed the expected Langmuir adsorption kinetics90 because of HIgG–GaHIgG binding.
Furthermore, to confirm the GaHIgG binding, fluorescence analysis was performed by observing samples under a microscope (Cal Zeiss, Axio Imager Z1) after incubation (Figure 3d). As observed from the figure, the accumulated fluorescence intensity becomes prominent, and it increased sequentially from a lower concentration to a higher concentration (0.5–50 μg/mL) of GaHIgG with respect to the blank sample. A similar variation in fluorescence intensity has been reported for FITC-labeled IgG antibody–antigen reactions.52,91,92
To illustrate the effectiveness in absorbance measurement, A540 = At (at time t = 10 min) and ΔAb were calculated using eq 2a. Figure 3e demonstrates the double Y axes graph (con. vs At along XY1 and con. vs ΔAb along XY2) of experimentally observed absorbance At and ΔAb. Both absorbances were found to be linearly proportional to the concentration of GaHIgG, which can explain the transport kinetics of molecular interactions.94,95 The change in At sometimes may give false results during nanoparticle immobilization, as well as when receptor binding was improper or any surface damage occurred during experimental procedures. However, the calculation of ΔAb by subtracting the buffer line after recording the absorbance response even for a lower concentration of target analytes precisely confirmed the binding and thus ensured the detection of target molecules.
The antibody–antigen complex molecules28,87 that have been captured on the surface due to binding can be described by a relation given in eq 2b when dissociation is negligible for 10 min of incubation where RHIgG(s) denotes the solid phase of the HIgG receptor immobilized on the surface, TGaHIgG(aq) is the GaHIgG target sample in aqueous medium, and RTHIgG–GaHIgG(s) is the immobilized HIgG–GaHIgG complex. Also, the rate [d(ΔAb)/dt] at which the LSPR absorption increases due to selective binding of GaHIgG can be calculated from eq 2c.
| 2b |
| 2c |
where Ka is the absorption rate, CR is the surface concentration of immobilized HIgG, CT is the concentration of GaHIgG target molecules, and CRT is the surface concentration of receptor–target complex molecules after binding of GaHIgG with HIgG.
The value of At is decided by the amount of GaHIgG immobilized on the surface, which was found to be around 10 times higher for a 50 μg/mL sample than for a 0.5 μg/mL sample. This indicates that Ka depends on the type of target molecules and the degree of affinity with its receptor as well as concentration.28,94
First, to understand the absorption rate d(ΔAb)/dt, all experimental curves were fitted within the linear region of GaHIgG association (Supporting Information S4, Figure S10). Then, all calculated slope values (t = 10 min of incubation) were further fitted linearly with concentration [CTvs d(ΔAb)/dt] according to eq 2c, and the binding constant was calculated from the slope of the graph (Supporting Information S4, Figure S11), which was found to be Ka = 1.83 × 10–5 (μg–1 mL s–1). A similar order of association rate constant was reported due to absorption of GaHIgG on the AuNP surface.93,94
Since this platform is based on RI sensing, the effective bulk RI of the surroundings of the nanoparticles is calculated using eq 1 after each process. The absolute RIs in each step for AuNPs, Cys, GA, HIgG, BSA, and GaHIgG at different concentrations were calculated and are presented in Table S1 (Supporting Information S4).
nt is defined as an effective RI of the surrounding medium of nanoparticles after 10 min of GaHIgG incubation (Figure 3b). It was found that the effective RI of the LSPR surface was modified from nbaseline = 1.36 (DI + PBS buffer line) to nt = 1.3648 for 50 μg/mL of the GaHIgG sample. Also, the change of RI Δnb[Δnb = nt – nbaseline] due to HIgG and GaHIgG binding was calculated using the calibration equation (eq 1) and is plotted in Figure 3f. It was observed that Δnb increased linearly (R2 = 0.98) when the GaHIgG concentration varied from 0.5 to 50 μg/mL. By observing the absorbance dynamics of the lowest sample concentration (0.5 μg/mL), the biosensing performance of this LSPR sensor was quantitatively evaluated and thus the LOD of the GaHIgG was analyzed. As seen in Figure 3c, there is a clear LSPR binding response for the 0.5 μg/mL sample above the buffer line (PBS + DI). Therefore, the LOD of GaHIgG for this sensor was found to be 0.5 μg/mL from this antibody–antigen study. Recently, the reported LOD of IgG in the PBS buffer was demonstrated between 0.1 to 0.2 μg/mL.90,95
Detection of the Plant Virus (ChiLCV)
After a successful demonstration of the ATR-LSPR platform as a potential biosensor, it was further employed to detect the plant virus, where ChiLCV DNA was chosen as the target molecule. The schematic of the nucleotide base pairs of target-specific viral ssDNA, receptor DNA, that is, the complementary DNA of the viral sample (cDNA), and non-specific ssDNA sample is presented in Figure 4a to get an idea of positive (true) and negative (false) detection of ChiLCV.
Figure 4.

Schematic representation of (a) nucleobase pair sequences of cDNA, viral DNA (ssDNA), and healthy DNA (non-specific ssDNA), (b) allowed conjugation for specific DNA binding, and (c) forbidden conjugation because of non-specific DNA binding with the cDNA-immobilized LSPR probe.
For positive detection, the probe was prepared with cDNA (5′ CATCAGAGCATTCTCACT 3′). The prepared LSPR probe will be allowed to conjugate only with viral ssDNA (3′ GTAGTCTCGTAAGAGTGA 5′), as depicted in Figure 4b (schematic). Whereas conjugation will be forbidden for the healthy leaf DNA sample or other viral DNA base pairs due to similar base pairs or non-specific base pairs and thus the detection will be negative for any non-specific DNA sample (Figure 4c). This specific and non-specific binding of DNAs will have an effect on absorbance dynamics, which confirms the detection of ChiLCV. In this detection process, the immobilized cDNA on the functionalized AuNPs having complementary bases with respect to ChiLCV DNA form hydrogen bonds during the pairing/hybridization.96−98 The absorbance A540 increased exponentially during this pairing. The amino acid sequence of this target ssDNA shows the maximum (96.1%) matching identity to its primer fragments.74 The conjugation initiated from the 5′ (sense) to the 3′ (anti-sense) direction and thus hybridization continued up to a maximum viral ssDNA sequence matching for 68–88% with cDNA sequence similarity.99
cDNA-Immobilized LSPR Probe
First, the cDNA (primer) binding on the LSPR surface was studied (Figure 5a). In this experiment, the nanoparticles and immobilization of cross-linkers Cys and GA were confirmed similarly by monitoring the LSPR absorbance, as explained in the previous sensogram (see GaHIgG detection, processes I, II, and III).
Figure 5.
(a) Time-resolved absorbance response steps during the cDNA (4.5 μg/mL) probe preparation on the AuNP probe surface, and the inset image shows the extended view of cDNA absorbance dynamics (starting point t = t3960 to t = t5340 for 23 min), (b) time-resolved absorbance response binding dynamics of four different concentrations of ChiLCV ssDNA (inset: extended view of the binding starting point t = t5590 to t = t6190 for 10 min), (c) double Y axes graph of viral ssDNA concentration vs observed normalized absorbance (con. vs ΔA540Norm along the left axis; XY1), and ssDNA concentration vs change in the RI Δn surrounding the medium of immobilized AuNPs was calculated after binding viral DNA (con. vs Δn).
Next, to prepare the receptor surface, the TE buffer (Tris–EDTA; 100 μM, pH 7.2) solution containing a short nucleotide of cDNA (40 μL, 4.5 μg/mL concentration) was introduced into the sample cavity. As seen in process IV, shown in the inset of Figure 5a, the cDNA exhibited an exponential increase in absorbance response due to GA-mediated interaction. The excess amount of cDNA was removed by rinsing with DI water. This interaction is known as Schiff base interaction between the aldehyde-functionalized surface97,98 and amine functional groups present in cNDA nucleotides. The cDNA covalently binds to amine-active sites on the ssDNA bases by hydrogen bonds100−102 and thus the cDNA-immobilized LSPR probe is prepared. The change in the RI during this cDNA probe preparation is given in Table S2 (Supporting Information S5). In the next step, to detect the viral ssDNA, the concentration of cDNA on the LPSR probe was maintained at 4.5 μg/mL.
Target Viral DNA Detection
Once the cDNA was immobilized on the surface, the ssDNA from the chilli plant infected with ChiLCV was introduced on the surface and the real-time change in the LSPR absorption dynamics was observed. First, the sample having 3.5 μg/mL concentration was introduced into the sample chamber and the change in absorbance was observed with time (Supporting Information S5, Figure S12).
Since the ssDNA binding mechanism depends on the homogeneous distribution of cDNA on the AuNP surface, the concentration of the ssDNA was varied from 0.5 to 3.5 μg/mL (in steps of 1.0 μg/mL) to find the binding limit (Figure 5b). The initial binding dynamics at lower concentrations did not have a clear exponential growth, but it follows good exponential behavior at higher concentrations. From this study, the normalized absorbance ΔA540Norm due to ssDNA binding was calculated (eq 1), which was found to vary linearly with the concentration (Figure 5c, along the left axis). For this absorbance, the corresponding RI change Δnb for ssDNA binding was calculated and found to vary from 2.4 × 10–4 to 8.7 × 10–4 RIU with the concentration (Figure 5c, along right axis), and the linear correlation between the concentration, RI, and absorbance of viral DNA was established. The change in RI during the concentration variation is listed in Table S2 (Supporting Information S5). Furthermore, the absorption rate or the hybridization rate of ssDNA was found using a similar concept as described above (eq 2c) and found to be Ka = 0.64 × 10–4 (μg–1 mL s–1).
The minimum detectable concentration of the ChiLCV ssDNA from the ATR-LSPR probe was found to be 0.5 μg/mL, and the LOD from Figure 5c was found to be at least 2 times higher, that is, 1.0 μg/mL, than the minimum detectable concentration.
A comparison in detection limit has been performed between the existing methods and the new ATR-LSPR method and is summarized in Table 1. As seen in the table, the PCR method is able to detect less than 1.5 ng/μL or (pM) concentration of a plant virus. This antibody test is very specific to a short segment of begomovirus nucleic acid, which prevents any false result and ensures quantitative standard detection. However, this method is costly, time-consuming, and laboratory-specific. Whereas, the ELISA method is a very specific antigen–antibody reaction, and the detection limit is about 10–100 ng/μL. The AuNP-based complementary oligonucleotide-immobilized probe technique using the ATR-based evanescent wave absorption method by monitoring LSPR has been studied to detect the begomovirus DNAs. This platform has potential for rapid, label-free, low-cost detection of plant pathogens for the development of plasmonic-based biosensors.
Table 1. Comparison of LOD Values between the Existing Methods and the New ATR-LSPR Method.
| ref no. | sensing method | receptor molecules | target molecules | sensitivity |
|---|---|---|---|---|
| (68) | PCR amplification | DNA A and DNA B genome | mungbean yellow mosaic India virus | 1.5–2.0 ng/μL |
| (72) | TAS-ELISAs | recombinant coat protein (CP) of tomato leaf curl virus | tomato yellow leaf curl virus | 0.25 μg/mL |
| (13) | ATR-FTIR using germanium crystal | biotin-coupled PE-BTN | streptavidin | 10 pM |
| (103) | ELISA | recombinant capsid CP of ChiVMV | chilli veinal mottle virus (ChiVMV) | 4.38–6.43 ng/μL |
| (87) | quartz crystal microgravimetry-based SPR | Staphylococcus aureus protein A and Streptococcus protein G | HIgG | 0.01–0.05 mg/mL |
| (20) | U-bent fiber optic probe | HIgG | GaHIgG | 0.1 μg/mL |
| (104) | gold and silver nanoparticle-based direct DNA hybridization assay-scanometric detection | thiol-modified oligonucleotide form CP gene | tomato leaf curl New Delhi virus ssDNA | 1 nM to 100 pM |
| in this work | AuNPs-based ART-LSPR | oligonucleotide from CP gene | ChiLCV ssDNA | 0.5 μg/mL |
Selectivity Study
After confirming the detection of ChiLCV, the specificity of the developed sensor was examined by using an unknown DNA sequence (non-specific DNA sample), which was derived from the healthy chilli leaf. The healthy leaf ssDNA binding test was studied on separate sensor probes after immobilizing cDNA on the functionalized AuNP surface. An aliquot of 1.0 μg/mL concentration of healthy DNA was used for this experiment. After incubating with healthy chilli ssDNA on the detector-linked surface, no prominent change in LSPR absorbance was observed, as shown in Figure 6 (olive green curve), compared to the lowest concentration (0.5 μg/mL) of viral DNA from the ChiLCV-infected leaf sample. The binding of ChiLCV-intercalated chilli leaf DNA with its probe cDNA is very specific because of the complementary nature of nucleobases. Other than this, the nucleotide sequence of any unknown DNAs could not initiate the hybridization process and hence no absorbance kinetics was observed for healthy leaf DNA. The time resolved absorbance response for allowed and forbidden conjugation is shown in Figure S13 (Supporting Information S5). A minor instant change in absorbance was observed, which was due to the analyte’s RI change. Hence, it is demonstrated that the developed sensor can detect the specific target plant viral pathogen.
Figure 6.

Real-time change in absorbance due to specific (viral ssDNA) binding and non-specific (healthy DNA) binding with cDNA-immobilized LSPR probes.
Conclusions
In this work, an ATR-LSPR-based optical platform was employed to demonstrate the detection of ChiLCV. It has been shown here that even with a small volume (20–40 μL) of sample, the LSPR absorbance signal could be monitored using this optical platform, so it has potential to be developed as a LSPR-based microdevice for biomedical applications. The LOD of this sensor was found to be 0.5 μg/mL for the IgG antigen–antibody-based interaction, which is the actual LOD of this LSPR probe. However, for the plant viral DNA sample, the LOD is 1.0 μg/mL where a distinct binding dynamics was observed as compared to non-specific DNA. Furthermore, the sensitivity of the proposed ATR-LSPR sensor could be improved by increasing the immobilization density of AuNPs. This biosensing platform could be used for the detection of other micro-organisms such as proteins, nucleic acids, viruses, bacteria, and fungi. A more comprehensive study is needed to identify other begomoviruses with a view of a control strategy for the management of economically important crop plants.
Materials
Gold (III) chloride trihydrate (30% wt of HAuCl4 in dilute hydrochloric acid), ys (HS–C2H4–NH2), APTES (99%), PBS, and Tris–EDTA (TE) buffer were purchased from Sigma-Aldrich. Trisodium citrate (Na3C6H11O10), acetic acid, and sucrose powder were procured from SD Fine-Chemicals Limited. The linkers GA (25% aqueous; OHC–CH2–CH2–CH2–CHO) and Cys dihydrochloride (96%; C4H12N2S2·2HCl) were bought from Merck Life Science Pvt. Ltd. The HIgG antibody, GaHIgG, and BSA were procured from Bangalore Genei Private Limited. Microscopic coverslips were purchased from Blue Star. All the solutions were prepared using DI water from the Milli-Q filtration system.
Acknowledgments
The authors would like to acknowledge the IRCC, SAIF, and IIT Bombay for allowing the usage of central facilities for characterization. Also, S.D. would like to thank the University Grants Commission (UGC), Government of India, for the research fellowship support. V.R.R. would like to thank the Science and Engineering Research Board, Government of India, for a J.C. Bose Fellowship.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01702.
Experimental details; functionalization of the substrate; synthesis of AuNPs; AuNP immobilization; antibody–antigen immobilization process; viral and non-viral sample preparation; RI solution preparation; HIgG–GaHIgG binding results; red shift in the LSPR peak; absorption rate calculation; RI calculation for IgG; schematic of allowed and forbidden conjugation of DNA; absorbance response of viral ssDNA; and RI calculation for viral samples (PDF)
Author Contributions
This manuscript was written through the contributions of all authors.
This work has been supported by funding from the Government of India for DST India Nano-Mission sponsored project (Development of Lab-on-chip platforms for Efficient and Automated Farming (LEAF); code: SR/NM/TP-56/2012).
The authors declare no competing financial interest.
Supplementary Material
References
- Tokel O.; Inci F.; Demirci U. Advances in plasmonic technologies for point of care applications. Chem. Rev. 2014, 114, 5728–5752. 10.1021/cr4000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman S. S.; McKeating K. S.; Cheng Q. Surface plasmon resonance: material and interface design for universal accessibility. Anal. Chem. 2018, 90, 19. 10.1021/acs.analchem.7b04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri M.; Bezaatpour A.; Jafari H.; Boukherroub R.; Szunerits S. Electrochemical methodologies for the detection of pathogens. ACS Sens. 2018, 3, 1069–1086. 10.1021/acssensors.8b00239. [DOI] [PubMed] [Google Scholar]
- McDonagh C.; Burke C. S.; MacCraith B. D. Optical chemical sensors. Chem. Rev. 2008, 108, 400–422. 10.1021/cr068102g. [DOI] [PubMed] [Google Scholar]
- Chandirasekar S.; Dharanivasan G.; Kasthuri J.; Kathiravan K.; Rajendiran N. Facile synthesis of bile salt encapsulated gold nanoparticles and its use in colorimetric detection of DNA. J. Phys. Chem. C 2011, 115, 15266–15273. 10.1021/jp2044465. [DOI] [Google Scholar]
- Khater M.; de la Escosura-Muñiz A.; Merkoçi A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. 10.1016/j.bios.2016.09.091. [DOI] [PubMed] [Google Scholar]
- Spackova B.; Wrobel P.; Bocková M.; Homola J. Optical biosensors based on plasmonic nanostructures: a review. Proc. IEEE 2016, 104, 2380–2408. 10.1109/jproc.2016.2624340. [DOI] [Google Scholar]
- Kurihara K.; Nakamura K.; Hirayama E.; Suzuki K. An absorption-based surface plasmon resonance sensor applied to sodium ion sensing based on an ion-selective optode membrane. Anal. Chem. 2002, 74, 6323–6333. 10.1021/ac0203241. [DOI] [PubMed] [Google Scholar]
- Sharma A. K.; Jha R.; Gupta B. D. Fiber-optic sensors based on surface plasmon resonance: a comprehensive review. IEEE Sens. J. 2007, 7, 1118–1129. 10.1109/jsen.2007.897946. [DOI] [Google Scholar]
- Tan A. J. Y.; Ng S. M.; Stoddart P. R.; Chua H. S. Trends and Applications of U-Shaped Fiber Optic Sensors: A Review. IEEE Sens. J. 2021, 21, 120–131. 10.1109/jsen.2020.3014190. [DOI] [Google Scholar]
- Fenzl C.; Hirsch T.; Baeumner A. J. Liposomes with high refractive index encapsulants as tunable signal amplification tools in surface plasmon resonance spectroscopy. Anal. Chem. 2015, 87, 11157–11163. 10.1021/acs.analchem.5b03405. [DOI] [PubMed] [Google Scholar]
- Mark S. S.; Sandhyarani N.; Zhu C.; Campagnolo C.; Batt C. A. Dendrimer-functionalized self-assembled monolayers as a surface plasmon resonance sensor surface. Langmuir 2004, 20, 6808–6817. 10.1021/la0495276. [DOI] [PubMed] [Google Scholar]
- Voue M.; Goormaghtigh E.; Homblé F.; Marchand-Brynaert J.; Conti J.; Devouge S.; De Coninck J. Biochemical interaction analysis on ATR devices: A wet chemistry approach for surface functionalization. Langmuir 2007, 23, 949–955. 10.1021/la061627j. [DOI] [PubMed] [Google Scholar]
- Li C.-T.; Chen H.-f.; Un I.-W.; Lee H.-C.; Yen T.-J. Study of optical phase transduction on localized surface plasmon resonance for ultrasensitive detection. Opt. Express 2012, 20, 3250–3260. 10.1364/oe.20.003250. [DOI] [PubMed] [Google Scholar]
- Chien F.-C.; Chen S.-J. A sensitivity comparison of optical biosensors based on four different surface plasmon resonance modes. Biosens. Bioelectron. 2004, 20, 633–642. 10.1016/j.bios.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Wolfbeis O. S. Fiber-optic chemical sensors and biosensors. Anal. Chem. 2008, 80, 4269–4283. 10.1021/ac800473b. [DOI] [PubMed] [Google Scholar]
- Sousa R. P. C. L.; Figueira R. B.; Costa S. P. G.; Raposo M. M. Optical Fiber Sensors for Biocide Monitoring: Examples, Transduction Materials, and Prospects. ACS Sens. 2020, 5, 3678–3709. 10.1021/acssensors.0c01615. [DOI] [PubMed] [Google Scholar]
- Wang X.-d.; Wolfbeis O. S. Fiber-Optic Chemical Sensors and Biosensors (2015-2019). Anal. Chem. 2019, 92, 397–430. 10.1021/acs.analchem.9b04708. [DOI] [PubMed] [Google Scholar]
- Kundu T.; Sai V. V. R.; Dutta R.; Titas S.; Kumar P.; Mukherjee S. Development of evanescent wave absorbance-based fibre-optic biosensor. Pramana 2010, 75, 1099–1113. 10.1007/s12043-010-0193-6. [DOI] [Google Scholar]
- Sai V. V. R.; Kundu T.; Mukherji S. Novel U-bent fiber optic probe for localized surface plasmon resonance based biosensor. Biosens. Bioelectron. 2009, 24, 2804–2809. 10.1016/j.bios.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Ekgasit S.; Thammacharoen C.; Knoll W. Surface plasmon resonance spectroscopy based on evanescent field treatment. Anal. Chem. 2004, 76, 561–568. 10.1021/ac035042v. [DOI] [PubMed] [Google Scholar]
- Kurihara K.; Suzuki K. Theoretical understanding of an absorption-based surface plasmon resonance sensor based on Kretchmann’s theory. Anal. Chem. 2002, 74, 696–701. 10.1021/ac010820+. [DOI] [PubMed] [Google Scholar]
- Lambert A. S.; Valiulis S. N.; Malinick A. S.; Tanabe I.; Cheng Q. Plasmonic biosensing with aluminum thin films under the Kretschmann configuration. Anal. Chem. 2020, 92, 8654–8659. 10.1021/acs.analchem.0c01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaera F. Probing liquid/solid interfaces at the molecular level. Chem. Rev. 2012, 112, 2920–2986. 10.1021/cr2002068. [DOI] [PubMed] [Google Scholar]
- Banas A.; Banas K.; Lo M. K. F.; Kansiz M.; Kalaiselvi S. M. P.; Lim S. K.; Loke J.; Breese M. B. H. Detection of High-Explosive Materials within Fingerprints by Means of Optical-Photothermal Infrared Spectromicroscopy. Anal. Chem. 2020, 92, 9649–9657. 10.1021/acs.analchem.0c00938. [DOI] [PubMed] [Google Scholar]
- Yang J.; Her J.-W. Purge-and-trap ATR/IR spectroscopic method for the detection of semivolatile aromatic compounds in soils. Anal. Chem. 1999, 71, 4690–4696. 10.1021/ac990180z. [DOI] [Google Scholar]
- Blond P.; Bevernaegie R.; Troian-Gautier L.; Lagrost C.; Hubert J.; Reniers F.; Raussens V.; Jabin I. Ready-to-use germanium surfaces for the development of FTIR-based biosensors for proteins. Langmuir 2020, 36, 12068–12076. 10.1021/acs.langmuir.0c02681. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Islam N.; Carbonell R. G.; Rojas O. J. Specific binding of immunoglobulin G with bioactive short peptides supported on antifouling copolymer layers for detection in quartz crystal microgravimetry and surface plasmon resonance. Anal. Chem. 2013, 85, 1106–1113. 10.1021/ac302874s. [DOI] [PubMed] [Google Scholar]
- Boulet-Audet M.; Byrne B.; Kazarian S. G. High-throughput thermal stability analysis of a monoclonal antibody by attenuated total reflection FT-IR spectroscopic imaging. Anal. Chem. 2014, 86, 9786–9793. 10.1021/ac502529q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiernan H.; Byrne B.; Kazarian S. G. Insight into heterogeneous distribution of protein aggregates at the surface layer using attenuated total reflection-Fourier transform infrared spectroscopic imaging. Anal. Chem. 2020, 92, 4760–4764. 10.1021/acs.analchem.0c00250. [DOI] [PubMed] [Google Scholar]
- Onodera K.; Hirano-Iwata A.; Miyamoto K.-i.; Kimura Y.; Kataoka M.; Shinohara Y.; Niwano M. Label-Free Detection of Protein–Protein Interactions at the GaAs/Water Interface through Surface Infrared Spectroscopy: Discrimination between Specific and Nonspecific Interactions by Using Secondary Structure Analysis. Langmuir 2007, 23, 12287–12292. 10.1021/la7022192. [DOI] [PubMed] [Google Scholar]
- Brewer S. H.; Anthireya S. J.; Lappi S. E.; Drapcho D. L.; Franzen S. Detection of DNA hybridization on gold surfaces by polarization modulation infrared reflection absorption spectroscopy. Langmuir 2002, 18, 4460–4464. 10.1021/la025613z. [DOI] [Google Scholar]
- Halpern A. R.; Chen Y.; Corn R. M.; Kim D. Surface plasmon resonance phase imaging measurements of patterned monolayers and DNA adsorption onto microarrays. Anal. Chem. 2011, 83, 2801–2806. 10.1021/ac200157p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma M.; Tawa K. Polydopamine thin films as protein linker layer for sensitive detection of interleukin-6 by surface plasmon enhanced fluorescence spectroscopy. ACS Appl. Mater. Interfaces 2016, 8, 22032–22038. 10.1021/acsami.6b06917. [DOI] [PubMed] [Google Scholar]
- Budde B.; Schartner J.; Tönges L.; Kötting C.; Nabers A.; Gerwert K. Reversible Immuno-Infrared Sensor for the Detection of Alzheimer’s Disease Related Biomarkers. ACS Sens. 2019, 4, 1851–1856. 10.1021/acssensors.9b00631. [DOI] [PubMed] [Google Scholar]
- Niegelhell K.; Leimgruber S.; Grießer T.; Brandl C.; Chernev B.; Schennach R.; Trimmel G.; Spirk S. Adsorption studies of organophosphonic acids on differently activated gold surfaces. Langmuir 2016, 32, 1550–1559. 10.1021/acs.langmuir.5b04467. [DOI] [PubMed] [Google Scholar]
- Touahir L.; Jenkins A. T. A.; Boukherroub R.; Gouget-Laemmel A. C.; Chazalviel J.-N.; Peretti J.; Ozanam F.; Szunerits S. Surface plasmon-enhanced fluorescence spectroscopy on silver based SPR substrates. J. Phys. Chem. C 2010, 114, 22582–22589. 10.1021/jp107402r. [DOI] [Google Scholar]
- Maalouli N.; Gouget-Laemmel A. C.; Pinchemel B.; Bouazaoui M.; Chazalviel J.-N.; Ozanam F.; Yang Y.; Burkhard P.; Boukherroub R.; Szunerits S. Development of a metal-chelated plasmonic interface for the linking of his-peptides with a droplet-based surface plasmon resonance read-off scheme. Langmuir 2011, 27, 5498–5505. 10.1021/la2005437. [DOI] [PubMed] [Google Scholar]
- Lin W.; Li Z. Detection and quantification of trace organic contaminants in water using the FT-IR-attenuated total reflectance technique. Anal. Chem. 2010, 82, 505–515. 10.1021/ac901192d. [DOI] [PubMed] [Google Scholar]
- De Alwis C.; Leftwich T. R.; Perrine K. A. New Approach to Simultaneous In Situ Measurements of the Air/Liquid/Solid Interface Using PM-IRRAS. Langmuir 2020, 36, 3404–3414. 10.1021/acs.langmuir.9b03958. [DOI] [PubMed] [Google Scholar]
- Perez-Guaita D.; Richardson Z.; Heraud P.; Wood B. Quantification and identification of microproteinuria using ultrafiltration and ATR-FTIR spectroscopy. Anal. Chem. 2020, 92, 2409–2416. 10.1021/acs.analchem.9b03081. [DOI] [PubMed] [Google Scholar]
- Giotta L.; Mastrogiacomo D.; Italiano F.; Milano F.; Agostiano A.; Nagy K.; Valli L.; Trotta M. Reversible binding of metal ions onto bacterial layers revealed by protonation-induced ATR-FTIR difference spectroscopy. Langmuir 2011, 27, 3762–3773. 10.1021/la104868m. [DOI] [PubMed] [Google Scholar]
- Cuba-Chiem L. T.; Huynh L.; Ralston J.; Beattie D. A. In situ particle film ATR FTIR spectroscopy of carboxymethyl cellulose adsorption on talc: binding mechanism, pH effects, and adsorption kinetics. Langmuir 2008, 24, 8036–8044. 10.1021/la800490t. [DOI] [PubMed] [Google Scholar]
- Wolfram H.; Wriedt T.. The Mie theory: basics and applications; Springer, 2012; Vol. 169, pp 60. [Google Scholar]
- Papoff F.; Hourahine B. Geometrical Mie theory for resonances in nanoparticles of any shape. Opt. Express 2011, 19, 21432–21444. 10.1364/oe.19.021432. [DOI] [PubMed] [Google Scholar]
- Yockell-Lelièvre H.; Lussier F.; Masson J.-F. Influence of the particle shape and density of self-assembled gold nanoparticle sensors on lspr and sers. J. Phys. Chem. C 2015, 119, 28577–28585. 10.1021/acs.jpcc.5b09570. [DOI] [Google Scholar]
- Zhang Z.; Chen Z.; Qu C.; Chen L. Highly sensitive visual detection of copper ions based on the shape-dependent LSPR spectroscopy of gold nanorods. Langmuir 2014, 30, 3625–3630. 10.1021/la500106a. [DOI] [PubMed] [Google Scholar]
- Shabaninezhad M.; Ramakrishna G. Theoretical investigation of size, shape, and aspect ratio effect on the LSPR sensitivity of hollow-gold nanoshells. J. Chem. Phys. 2019, 150, 144116. 10.1063/1.5090885. [DOI] [PubMed] [Google Scholar]
- Nehl C. L.; Hafner J. H. Shape-dependent plasmon resonances of gold nanoparticles. J. Mater. Chem. 2008, 18, 2415–2419. 10.1039/b714950f. [DOI] [Google Scholar]
- Jiang G.; Baba A.; Ikarashi H.; Xu R.; Locklin J.; Kashif K. R.; Shinbo K.; Kato K.; Kaneko F.; Advincula R. Signal enhancement and tuning of surface plasmon resonance in Au nanoparticle/polyelectrolyte ultrathin films. J. Phys. Chem. C 2007, 111, 18687–18694. 10.1021/jp075986e. [DOI] [Google Scholar]
- Zhang Z.; Wang H.; Chen Z.; Wang X.; Choo J.; Chen L. Plasmonic colorimetric sensors based on etching and growth of noble metal nanoparticles: Strategies and applications. Biosens. Bioelectron. 2018, 114, 52–65. 10.1016/j.bios.2018.05.015. [DOI] [PubMed] [Google Scholar]
- Hutter E.; Cha S.; Liu J.-F.; Park J.; Yi J.; Fendler J. H.; Roy D. Role of substrate metal in gold nanoparticle enhanced surface plasmon resonance imaging. J. Phys. Chem. B 2001, 105, 8–12. 10.1021/jp003565q. [DOI] [Google Scholar]
- Ghosh H.; Bürgi T. Adsorption of Gold and Silver Nanoparticles on Polyelectrolyte Layers and Growth of Polyelectrolyte Multilayers: An In Situ ATR-IR Study. J. Phys. Chem. C 2013, 117, 26652–26658. 10.1021/jp409241t. [DOI] [Google Scholar]
- Srisombat L.-o.; Park J.-S.; Zhang S.; Lee T. R. Preparation, characterization, and chemical stability of gold nanoparticles coated with mono-, bis-, and tris-chelating alkanethiols. Langmuir 2008, 24, 7750–7754. 10.1021/la800511g. [DOI] [PubMed] [Google Scholar]
- Volkert A. A.; Subramaniam V.; Ivanov M. R.; Goodman A. M.; Haes A. J. Salt-mediated self-assembly of thioctic acid on gold nanoparticles. ACS Nano 2011, 5, 4570–4580. 10.1021/nn200276a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F.; Dahl G. T.; Besztejan S.; Schroer M. A.; Lehmkühler F.; Grübel G.; Vossmeyer T.; Lange H. Ligand layer engineering to control stability and interfacial properties of nanoparticles. Langmuir 2016, 32, 7897–7907. 10.1021/acs.langmuir.6b01704. [DOI] [PubMed] [Google Scholar]
- Gupta A.; Moyano D. F.; Parnsubsakul A.; Papadopoulos A.; Wang L.-S.; Landis R. F.; Das R.; Rotello V. M. Ultrastable and biofunctionalizable gold nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 14096–14101. 10.1021/acsami.6b02548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J. H.; Kim K.-I.; Cho H. H.; Lee J. W.; Lee B. S.; Yoon S.; Park K. J.; Lee S.; Kim J.; Whang D.; Lee J. H. Ultrastable-stealth large gold nanoparticles with DNA directed biological functionality. Langmuir 2015, 31, 13773–13782. 10.1021/acs.langmuir.5b03534. [DOI] [PubMed] [Google Scholar]
- Tan Y. N.; Lee K. H.; Su X. Study of Single-Stranded DNA Binding Protein-Nucleic Acids Interactions using Unmodified Gold Nanoparticles and Its Application for Detection of Single Nucleotide Polymorphisms. Anal. Chem. 2011, 83, 4251–4257. 10.1021/ac200525a. [DOI] [PubMed] [Google Scholar]
- Chen H.; Park S.-G.; Choi N.; Moon J.-I.; Dang H.; Das A.; Lee S.; Kim D.-G.; Chen L.; Choo J. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020, 167, 112496. 10.1016/j.bios.2020.112496. [DOI] [PubMed] [Google Scholar]
- Chowdhury A. D.; Takemura K.; Khorish I. M.; Nasrin F.; Ngwe Tun M. M.; Morita K.; Park E. Y. The detection and identification of dengue virus serotypes with quantum dot and AuNP regulated localized surface plasmon resonance. Nanoscale Adv. 2020, 2, 699–709. 10.1039/c9na00763f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Chen C.; Qian M.; Zhao X. S. Aptamer biosensor for protein detection using gold nanoparticles. Anal. Biochem. 2008, 373, 213–219. 10.1016/j.ab.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Ragsdale N. N.; Siegel M. R. New approaches to chemical control of plant pathogens. ACS Symp. Ser. 1985, 276, 35–45. 10.1021/bk-1985-0276.ch003. [DOI] [Google Scholar]
- Horká M.; Horký J.; Matoušková H.; Šlais K. Separation of plant pathogens from different hosts and tissues by capillary electromigration techniques. Anal. Chem. 2007, 79, 9539–9546. 10.1021/ac701718v. [DOI] [PubMed] [Google Scholar]
- Adamson H.; Ajayi M. O.; Campbell E.; Brachi E.; Tiede C.; Tang A. A.; Adams T. L.; Ford R.; Davidson A.; Johnson M.; McPherson M. J.; Tomlinson D. C.; Jeuken L. J. C. Affimer-Enzyme-Inhibitor Switch Sensor for Rapid Wash-free Assays of Multimeric Proteins. ACS Sens. 2019, 4, 3014–3022. 10.1021/acssensors.9b01574. [DOI] [PubMed] [Google Scholar]
- Akhter M. S.; Akanda A. M.; Kobayashi K.; Jain R. K.; Mandal B. Plant virus diseases and their management in Bangladesh. Crop Prot. 2019, 118, 57–65. 10.1016/j.cropro.2018.11.023. [DOI] [Google Scholar]
- Bikash M.; Rao G. P.; Baranwal V. K.; Jain R. K.. A Century of Plant Virology in India; Springer, 2017, pp 76. [Google Scholar]
- Rouhibakhsh A.; Priya J.; Periasamy M.; Haq Q. M. I.; Malathi V. G. An improved DNA isolation method and PCR protocol for efficient detection of multicomponents of begomovirus in legumes. J. Virol. Methods 2008, 147, 37–42. 10.1016/j.jviromet.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Verma S.; Saxena S.. Recent advancement in diagnosis of Begomoviruses. Begomoviruses: Occurrence and Management in Asia and Africa; Springer, 2017, pp 33–50. [Google Scholar]
- Singh S.; Nirmalkar V. K.; Awasthi L. P.. Recent advances in begomovirus research in India. Applied Plant Virology; Academic Press, 2020; pp 493–513. [Google Scholar]
- Nirbhay K.; Singh A. K.; Brotati C.; Supriya C. Recent advances in geminivirus detection and future perspectives. J. Plant Prot. Sci. 2010, 2, 1–18. [Google Scholar]
- Seepiban C.; Charoenvilaisiri S.; Warin N.; Bhunchoth A.; Phironrit N.; Phuangrat B.; Chatchawankanphanich O.; Attathom S.; Gajanandana O. Development and application of triple antibody sandwich enzyme-linked immunosorbent assays for begomovirus detection using monoclonal antibodies against Tomato yellow leaf curl Thailand virus. Virol. J. 2017, 14, 99. 10.1186/s12985-017-0763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.; Dixit C. K.; Woolley R.; O’Kennedy R.; McDonagh C. Label-Free Optical Characterization Methods for Detecting Amine Silanization-Driven Gold Nanoparticle Self-Assembly. Langmuir 2011, 27, 10421–10428. 10.1021/la202364c. [DOI] [PubMed] [Google Scholar]
- Senanayake D. M. J. B.; Varma A.; Mandal B. Virus-vector Relationships, Host Range, Detection and Sequence Comparison of Chilli leaf curl virus Associated with an Epidemic of Leaf Curl Disease of Chilli in Jodhpur, India. J. Phytopathol. 2012, 160, 146–155. 10.1111/j.1439-0434.2011.01876.x. [DOI] [Google Scholar]
- Dinda E.; Rashid M. H.; Biswas M.; Mandal T. K. Redox-active ionic-liquid-assisted one-step general method for preparing gold nanoparticle thin films: applications in refractive index sensing and catalysis. Langmuir 2010, 26, 17568–17580. 10.1021/la103084t. [DOI] [PubMed] [Google Scholar]
- Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- Kanwa N.; Patnaik A.; De S. K.; Ahamed M.; Chakraborty A. Effect of Surface Ligand and Temperature on Lipid Vesicle-Gold Nanoparticle Interaction: A Spectroscopic Investigation. Langmuir 2019, 35, 1008–1020. 10.1021/acs.langmuir.8b03673. [DOI] [PubMed] [Google Scholar]
- Browning-Kelley M. E.; Wadu-Mesthrige K.; Hari V.; Liu G. Y. Atomic force microscopic study of specific antigen/antibody binding. Langmuir 1997, 13, 343–350. 10.1021/la960918x. [DOI] [Google Scholar]
- Sorci M.; Dassa B.; Liu H.; Anand G.; Dutta A. K.; Pietrokovski S.; Belfort M.; Belfort G. Oriented covalent immobilization of antibodies for measurement of intermolecular binding forces between zipper-like contact surfaces of split inteins. Anal. Chem. 2013, 85, 6080–6088. 10.1021/ac400949t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Salmain M.; Liedberg B.; Boujday S. Naked Eye Immunosensing of Food Biotoxins Using Gold Nanoparticle-Antibody Bioconjugates. ACS Appl. Nano Mater. 2019, 2, 4150–4158. 10.1021/acsanm.9b00598. [DOI] [Google Scholar]
- Batalla P.; Fuentes M.; Mateo C.; Grazu V.; Fernandez-Lafuente R.; Guisan J. M. Covalent immobilization of antibodies on finally inert support surfaces through their surface regions having the highest densities in carboxyl groups. Biomacromolecules 2008, 9, 2230–2236. 10.1021/bm8003594. [DOI] [PubMed] [Google Scholar]
- Sadik O. A.; Xu H.; Gheorghiu E.; Andreescu D.; Balut C.; Gheorghiu M.; Bratu D. Differential Impedance Spectroscopy for Monitoring Protein Immobilization and Antibody–Antigen Reactions. Anal. Chem. 2002, 74, 3142–3150. 10.1021/ac0156722. [DOI] [PubMed] [Google Scholar]
- Tumolo T.; Angnes L.; Baptista M. S. Determination of the refractive index increment (dn/dc) of molecule and macromolecule solutions by surface plasmon resonance. Anal. Biochem. 2004, 333, 273–279. 10.1016/j.ab.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Kabiraz D. C.; Morita K.; Sakamoto K.; Kawaguchi T. Mechanism of surface plasmon resonance sensing by indirect competitive inhibition immunoassay using Au nanoparticle labeled antibody. Talanta 2017, 172, 1–7. 10.1016/j.talanta.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Höbel S.; Vornicescu D.; Bauer M.; Fischer D.; Keusgen M.; Aigner A. A novel method for the assessment of targeted PEI-based nanoparticle binding based on a static surface plasmon resonance system. Anal. Chem. 2014, 86, 6827–6835. 10.1021/ac402001q. [DOI] [PubMed] [Google Scholar]
- Islam N.; Gurgel P. V.; Rojas O. J.; Carbonell R. G. Effects of Composition of Oligo(ethylene glycol)-Based Mixed Monolayers on Peptide Grafting and Human Immunoglobulin Detection. J. Phys. Chem. C 2014, 118, 5361–5373. 10.1021/jp411469u. [DOI] [Google Scholar]
- Islam N.; Gurgel P. V.; Rojas O. J.; Carbonell R. G. Use of a Branched Linker for Enhanced Biosensing Properties in IgG Detection from Mixed Chinese Hamster Ovary Cell Cultures. Bioconjugate Chem. 2019, 30, 815–825. 10.1021/acs.bioconjchem.8b00918. [DOI] [PubMed] [Google Scholar]
- Evans E.; Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. 10.1016/s0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanov I.; Fiorucci S.; Tennikova T. Receptor-ligand interactions: Advanced biomedical applications. Mater. Sci. Eng. C 2016, 68, 890–903. 10.1016/j.msec.2016.07.072. [DOI] [PubMed] [Google Scholar]
- Hu W.; Lu Z.; Liu Y.; Li C. M. In situ surface plasmon resonance investigation of the assembly process of multiwalled carbon nanotubes on an alkanethiol self-assembled monolayer for efficient protein immobilization and detection. Langmuir 2010, 26, 8386–8391. 10.1021/la9048105. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T.; Shirdel B.; Mansoori B.; Baradaran B. Dual sensitivity enhancement in gold nanoparticle-based lateral flow immunoassay for visual detection of carcinoembryonic antigen. Anal. Sci. Adv. 2020, 1, 161–172. 10.1002/ansa.202000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran Q.H.; Mai A.T.; Nguyen T.T.; Nguyen T.H.H. Towards the use of protein A-tagged gold nanoparticles for signal amplification of electrochemical immunosensors in virus detection. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2012, 3, 025013. 10.1088/2043-6262/3/2/025013. [DOI] [Google Scholar]
- Kapil M. A.; Herr A. E. Binding kinetic rates measured via electrophoretic band crossing in a pseudohomogeneous format. Anal. Chem. 2014, 86, 2601–2609. 10.1021/ac403829z. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Carbonell R. G.; Rojas O. J. Bioactive cellulose nanofibrils for specific human IgG binding. Biomacromolecules 2013, 14, 4161–4168. 10.1021/bm4007979. [DOI] [PubMed] [Google Scholar]
- Yin Z.; Guo H.; Li Y.; Chiu J.; Tian L. Ultrastable Plasmonic Bioink for Printable Point-Of-Care Biosensors. ACS Appl. Mater. Interfaces 2020, 12, 35977–35985. 10.1021/acsami.0c11799. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Li R.; Ba S.; Lu Z.; Pitsinos E. N.; Li T.; Nicolaou K. C. DNA Binding and Cleavage Modes of Shishijimicin A. J. Am. Chem. Soc. 2019, 141, 7842–7852. 10.1021/jacs.9b01800. [DOI] [PubMed] [Google Scholar]
- Jain M. L.; Bruice P. Y.; Szabó I. E.; Bruice T. C. Incorporation of positively charged linkages into DNA and RNA backbones: a novel strategy for antigene and antisense agents. Chem. Rev. 2012, 112, 1284–1309. 10.1021/cr1004265. [DOI] [PubMed] [Google Scholar]
- Sobczyk L.; Grabowski S. J.; Krygowski T. M. Interrelation between H-bond and Pi-electron delocalization. Chem. Rev. 2005, 105, 3513–3560. 10.1021/cr030083c. [DOI] [PubMed] [Google Scholar]
- Ebner A.; Wildling L.; Kamruzzahan A. S. M.; Rankl C.; Wruss J.; Hahn C. D.; Hölzl M.; Zhu R.; Kienberger F.; Blaas D.; Hinterdorfer P.; Gruber H. J. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjugate Chem. 2007, 18, 1176–1184. 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- Jia Y.; Li J. Molecular assembly of Schiff base interactions: construction and application. Chem. Rev. 2015, 115, 1597–1621. 10.1021/cr400559g. [DOI] [PubMed] [Google Scholar]
- Wolter F. E.; Schneider K.; Davies B. P.; Socher E. R.; Nicholson G.; Seitz O.; Süssmuth R. D. Total Synthesis of Proximicin A–C and Synthesis of New Furan-Based DNA Binding Agents. Org. Lett. 2009, 11, 2804–2807. 10.1021/ol901003p. [DOI] [PubMed] [Google Scholar]
- Shi K.; Carpenter M. A.; Banerjee S.; Shaban N. M.; Kurahashi K.; Salamango D. J.; McCann J. L.; Starrett G. J.; Duffy J. V.; Demir Ö.; Amaro R. E.; Harki D. A.; Harris R. S.; Aihara H. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Biol. 2017, 24, 131. 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlermroj R.; Himananto O.; Seepiban C.; Kumpoosiri M.; Warin N.; Gajanandana O.; Elliott C. T.; Karoonuthaisiri N. Antibody array in a multiwell plate format for the sensitive and multiplexed detection of important plant pathogens. Anal. Chem. 2014, 86, 7049–7056. 10.1021/ac501424k. [DOI] [PubMed] [Google Scholar]
- Dharanivasan G.; Jesse D. M. I.; Muthuramalingam T. R.; Rajendran G.; Shanthi S.; Kathiravan K. Scanometric Detection of Tomato Leaf Curl New Delhi Viral DNA Using Mono- and Bifunctional AuNP-Conjugated Oligonucleotide Probes. ACS Omega 2019, 4, 10094–10107. 10.1021/acsomega.9b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






