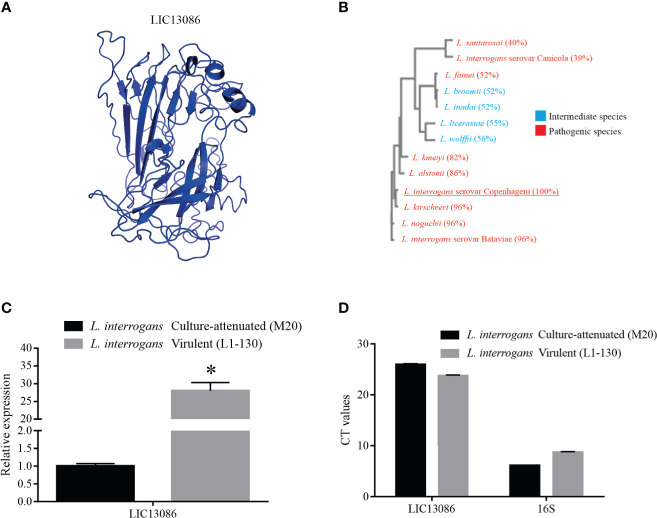
Figure 1.
In silico analysis of LIC13086 and real-time PCR native expression quantification. (A) Tertiary model of LIC13086 generated by molecular modeling in the I-TASSER webserver and visualized by PyMoL. (B) Dendogram resulting of multiple alignments performed by Clustal Omega of LIC13086 with other similar sequences of different species of Leptospira identified by BLASTp (pathogenic in red; intermediate in blue). (C) LIC13086 relative gene expression in culture-attenuated and virulent strains of L. interrogans by qPCR. Gene expression was normalized to 16S gene. (D) CT values of mRNA expression in culture-attenuated and virulent strains of L. interrogans. The bars represent the means of three different experiments in triplicates with the corresponding standard deviation. *Indicates significant difference calculated by Student t-test with p-values < 0.05.