Figure 2. PrimPol is rapidly recruited to ICLs in an RPA‐dependent manner.
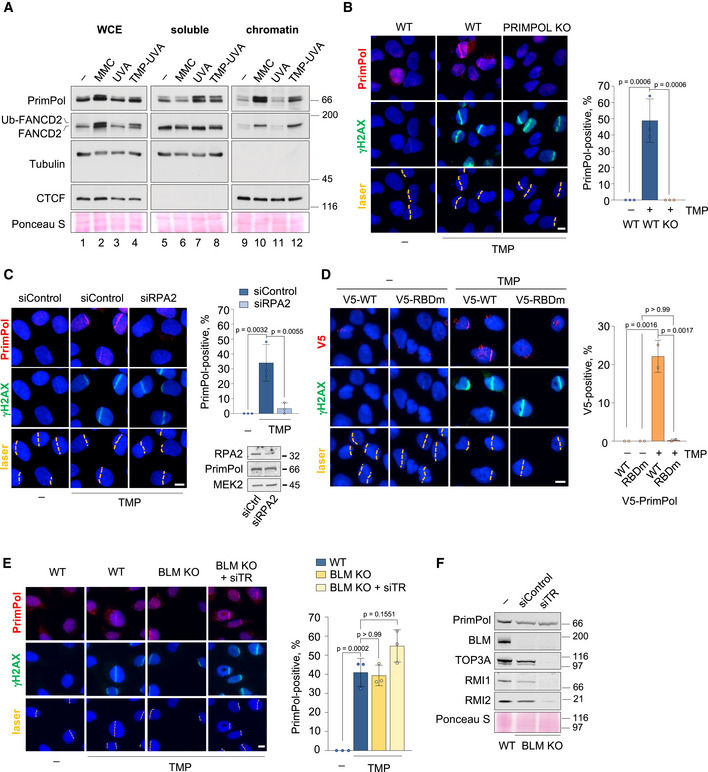
- Immunoblots show the levels of the indicated proteins in whole cell extracts (WCE), soluble and chromatin‐enriched fractions. Tubulin and CTCF are shown as controls for soluble and chromatin‐bound proteins, respectively. The position of the ubiquitylated form of FANCD2 (Ub‐FANCD2) is indicated. Ponceau S staining is shown as a loading control.
- Confocal microscopy images of PrimPol and γH2AX (damage control) immunofluorescence staining in control (UVA) or TMP‐UVA‐laser irradiated (10 µM TMP, 2 h followed by UVA laser irradiation) WT and PRIMPOL KO cells. Nuclear DNA is counterstained with DAPI. UVA laser path is indicated. Scale bar, 10 µm. Histograms (right panel) show the average percentage of PrimPol‐positive cells and SD of three assays (n ≥ 100 cells per TMP‐treated conditions in each replicate). Circle dots in each column represent the values of individual replicates. Statistical analysis was conducted with one‐way ANOVA followed by Bonferroni post‐test. P‐values of individual comparisons are indicated.
- Confocal microscopy images of PrimPol and γH2AX (damage control) immunofluorescence staining in control (UVA) or TMP‐UVA‐treated cells following control or RPA2 downregulation. Nuclear DNA is counterstained with DAPI. UVA laser path is indicated. Scale bar, 10 µm. Histograms (right panel) show the average PrimPol‐positive cells and SD of three assays (n ≥ 100 cells per TMP‐treated conditions in each replicate). Circle dots in each column represent the values of individual replicates. Statistical analysis was conducted with one‐way ANOVA followed by Bonferroni post‐test. P‐values of individual comparisons are indicated.
- Confocal microscopy images of PrimPol and γH2AX (damage control) immunofluorescence staining in control (UVA) or TMP‐UVA‐treated PRIMPOL KO cells stably expressing V5‐tagged WT or RPA binding domain mutant (RBDm) PrimPol versions. Nuclear DNA is counterstained with DAPI. UVA laser path is indicated. Scale bar, 10 µm. Histograms (right panel) show the average percentage of PrimPol‐positive cells and SD of two assays (n ≥ 100 cells per condition in each replicate). Circle dots in each column represent the values of individual replicates. Statistical analysis was conducted with one‐way ANOVA followed by Bonferroni post‐test. P‐values of individual comparisons are indicated.
- Confocal microscopy images of PrimPol and γH2AX (damage control) immunofluorescence staining in control (UVA) or TMP‐UVA‐treated WT and BLM KO cells upon downregulation of TOP3A, RMI1, and RMI2 (siTR) when indicated. Nuclear DNA is counterstained with DAPI. UVA laser path is indicated. Scale bar, 10 µm. Histograms (right panel) show the average percentage of PrimPol‐positive cells and SD of three assays (n ≥ 250 cells per condition). Circle dots in each column represent the values of individual replicates. Statistical analysis was conducted with one‐way ANOVA followed by Bonferroni post‐test.
- Immunoblots show the levels of BLM, TOP3A, RMI1, RMI2, and PrimPol after TOP3A, RMI1, and RMI2 downregulation in WT and BLM KO cells. Ponceau S is shown as loading control.
Source data are available online for this figure.
