Figure EV5. (related to Fig 5). Delayed clearance of FANCD2 mono‐ubiquitylation in PRIMPOL KO cells.
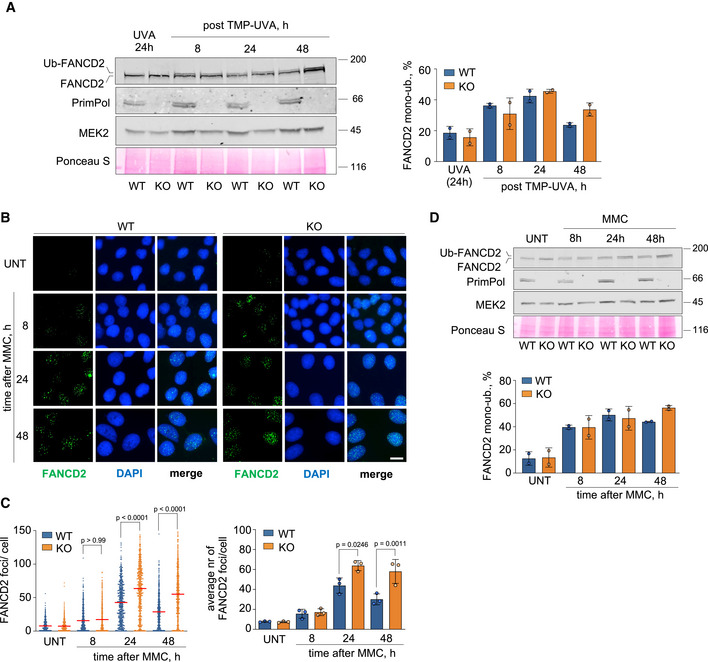
- Immunoblots showing the ubiquitylation of FANCD2 and the presence of PrimPol in WT and PRIMPOL KO cells at the indicated time points after treatment with TMP‐UVA (2 µM TMP for 2 h followed by 10 s irradiation). MEK2 and Ponceau S are shown as loading controls. To evaluate FANCD2 monoubiquitylation, the intensity of the upper and lower bands was calculated with ImageJ. The intensity of the top band was divided by the sum of upper and lower bands. Histograms (right panel) represent the percentage of FANCD2 mono‐ubiquitylation in each condition (average and SD of two replicates). Circle dots in each column represent the values of individual replicates.
- Confocal IF microscopy images of FANCD2 staining at the indicated times in control or MMC‐treated (0.5 µg/ml; 2 h) WT and PRIMPOL KO cells. Nuclear DNA is counterstained with DAPI. Scale bar, 25 µm.
- Dot plot (left panel) shows the distribution and mean (horizontal red line) of FANCD2 foci number per cell in each condition. The pool of three replicates is represented (n ≥ 100 cells per condition). Statistical analysis was conducted with Kruskal–Wallis test and Dunn’s post‐test. P‐values of the indicated comparisons are shown. Histograms (right panel) show the average foci number and SD of three assays in each case. Circle dots in each column represent the values of individual replicates. Statistical analysis was conducted with one‐way ANOVA followed by Bonferroni post‐test. P‐values are indicated.
- Immunoblots showing the levels and ubiquitylation of FANCD2 and the presence of PrimPol in WT and PRIMPOL KO cells at the indicated time points after treatment with 0,5 µg/ml MMC (2 h). MEK2 and Ponceau S are shown as loading controls. Histograms (bottom panel) represent the percentage of FANCD2 mono‐ubiquitylation in each condition (average and SD of two replicates), calculated as in A. Circle dots in each column represent the values of individual replicates.
Source data are available online for this figure.
