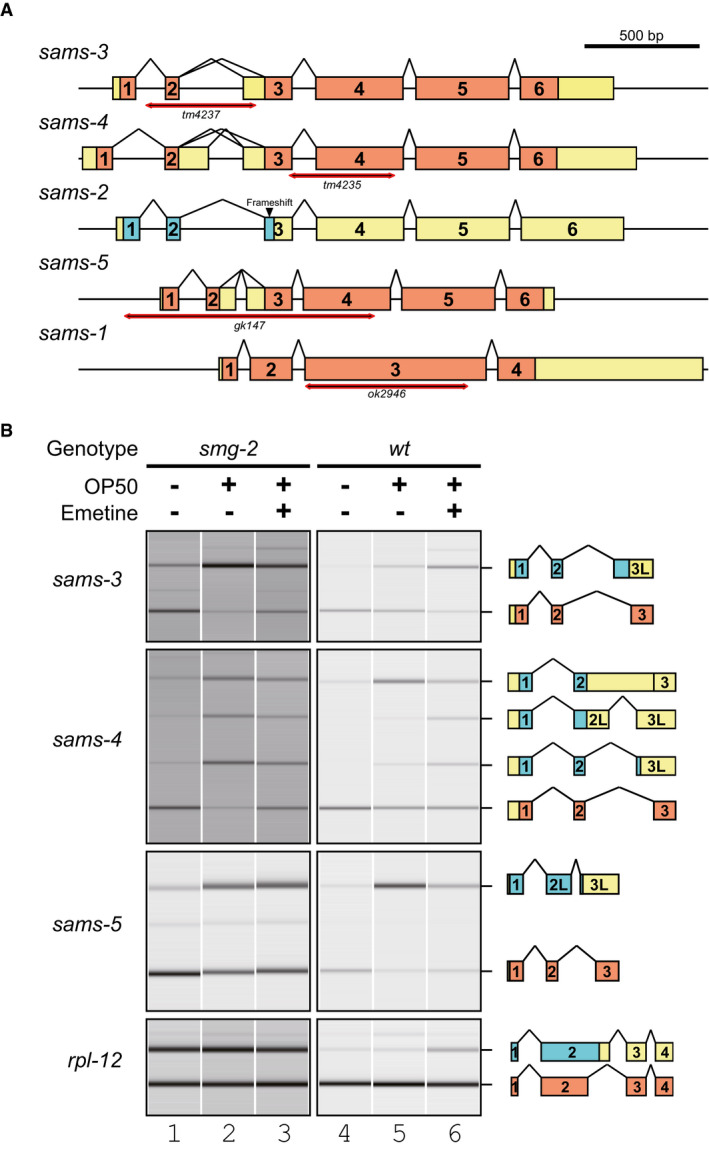
BAlternative splicing of sams‐3, sams‐4, and sams‐5 in unfed and fed worms without or with a protein synthesis inhibitor emetine. Synchronized L1 larvae of an NMD‐deficient strain KH1668: smg‐2
(yb979) (lanes 1‐3) and a wild‐type strain N2 (lanes 4‐6) were incubated in S‐complete medium alone (lanes 1, 4), with a standard E. coli strain OP50 (OD600 = 10.0) (lanes 2, 5) or OP50 supplemented with 10 mg/ml emetine (lanes 3, 6) for 3 h at 20°C. Total RNAs were extracted from whole animals and subjected to semi‐quantitative RT–PCR, whose products were analyzed by capillary electrophoresis. Representative gel‐like presentation is indicated (n = 3). Schematic structure of each PCR product is indicated on the right. Open reading frames (ORFs) for full‐length and truncated proteins are in orange and cyan, respectively. rpl‐12 was used as an unaffected control.