Figure EV2. Nucleotide sequence alignment of genomic regions spanning from exon 2 through exon 3 of the sams genes in C. elegans .
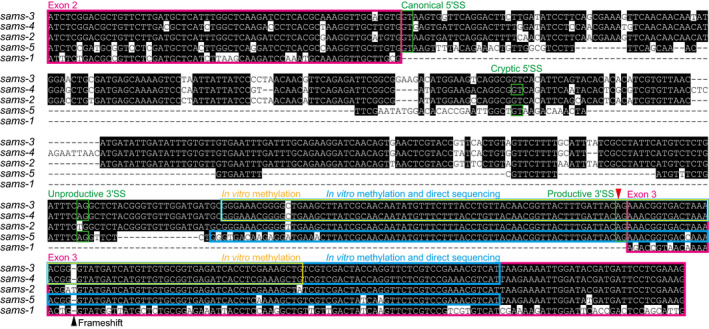
The sequences were aligned with Clustal V algorithm followed by manual adjustment. Conserved nucleotides are highlighted in black. Exons are boxed in magenta. Canonical and cryptic 5′SSs as well as proximal/unproductive and distal/productive 3′SSs are boxed in green. A yellow box indicates a sams‐3/sams‐4 RNA fragment used in in␣vitro methylation analysis with recombinant METT‐10 (Fig 5). Cyan boxes indicate sams‐3/sams‐4 and sams‐5 RNA fragments used in Nanopore direct sequencing after in␣vitro methylation (Fig 6). A red arrowhead indicates the m6A modification site. A black arrowhead indicates a frame‐shifting insertion in sams‐2. Note that sams‐1 lacks intron 2.
