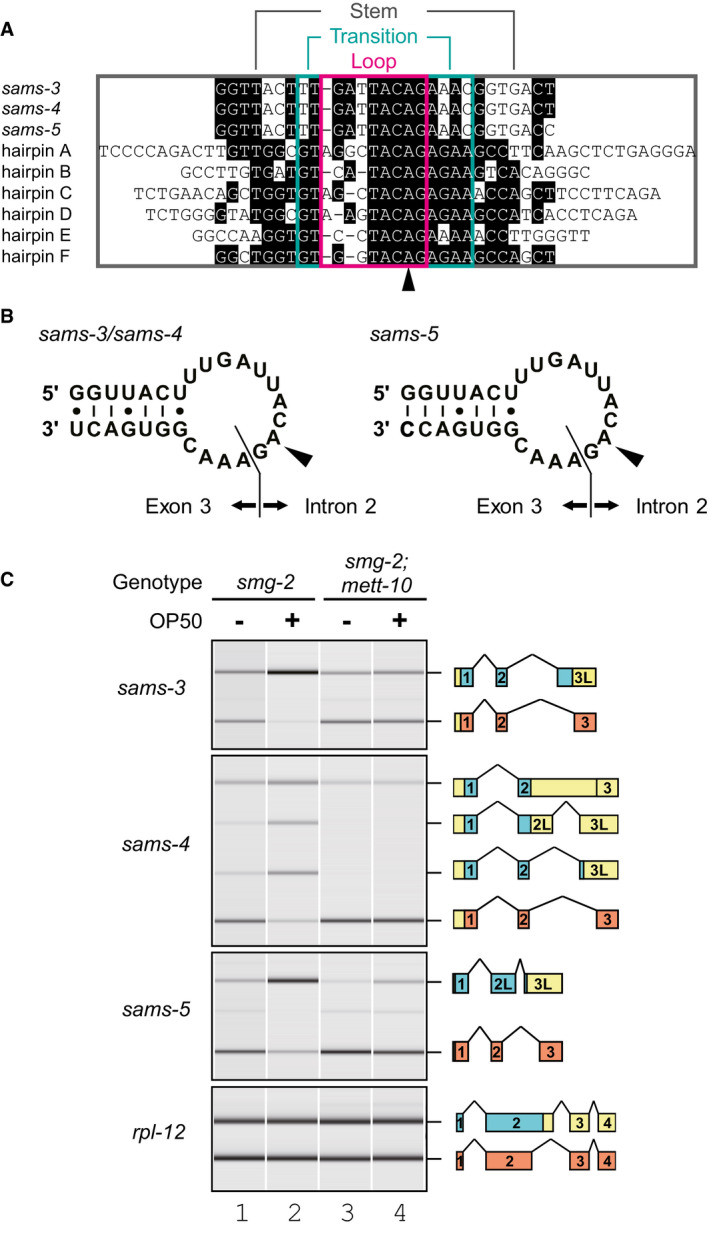
Figure 4. mett‐10 is required for effective production of NMD isoforms from the sams genes in␣vivo .
-
ANucleotide sequence alignment of putative stem‐loop structures in the C. elegans sams‐3, sams‐4, and sams‐5 genes together with six hairpin structures in 3′UTR of the human MAT2A gene. Conserved residues are shaded in black. Loop, transition, and stem regions of the MAT2A hairpins (Doxtader et␣al, 2018) are boxed in magenta, green, and gray, respectively. An arrowhead indicates adenine bases in the MAT2A hairpins that are specifically modified into m6A by METTL16 in␣vitro (Shima et␣al, 2017).
-
BSchematic representation of the predicted hairpin structures in sams‐3/sams‐4 (left) and sams‐5 (right) pre‐mRNAs. The boundaries between intron 2 and exon 3 are indicated. Arrowheads indicate the m6A modification sites.
-
CAlternative splicing of sams‐3, sams‐4, and sams‐5 in smg‐2 (yb979) and smg‐2 (yb979); mett‐10 (ok2204) mutants. Synchronized L1 larvae of each strain were incubated in S‐complete medium alone (lanes 1 and 3) or with OP50 (lanes 2 and 4) for 3 h at 20°C, and the splicing patterns were analyzed and presented as in Fig 2B.
