Figure EV5. Direct RNA sequencing with Nanopore technologies coupled with machine learning‐based classification reveals m6A modification of endogenous sams mRNAs.
-
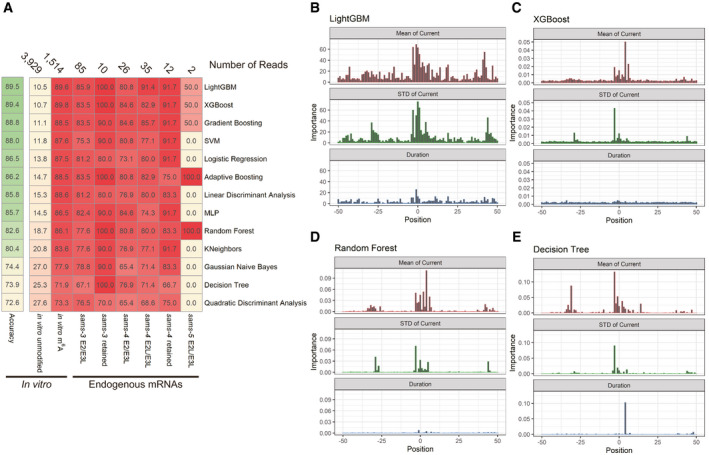
ASummary of machine learning‐based classification of sams mRNA reads in Nanopore direct RNA sequencing data with thirteen algorithms. Machine learning was conducted, and the results are shown as in Fig 6E. Numbers of reads used in the tests are indicated at the top; percentages of reads classified as “m6A‐modified” are shown with color code.
-
B–ERelative importance of the mean (top), standard deviation (middle), and duration time (bottom) of normalized Nanopore currents at nucleotide positions −50 through +50 relevant to the m6A site for LightGBM (B), XGBoost (C), Random Forest (D), and Decision Tree (E).
