Figure 6. Endogenous unproductive sams mRNAs have m6A modification at the AG dinucleotide of the distal 3'SS.
-
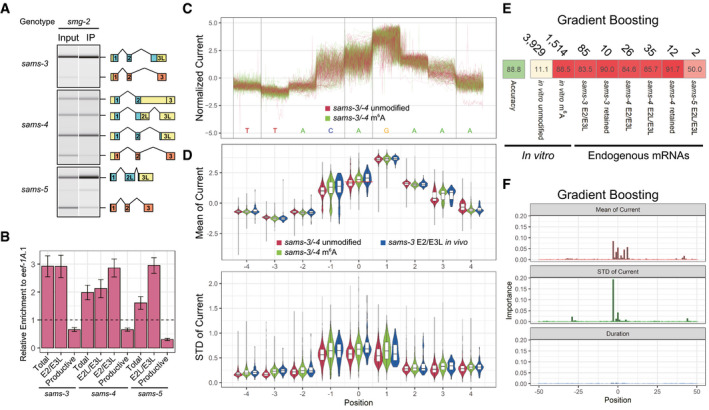
A, Bm6A‐IP specifically enriched sams mRNA isoforms that retain the AG dinucleotide at the distal/productive 3′SS. (A) Representative gel‐like images (n = 3) of semi‐quantitative RT–PCR analysis of sams‐3, sams‐4, and sams‐5 in input and immunoprecipitated (IP) RNAs from the smg‐2 (yb979) mutant. The splicing patterns were analyzed and presented as in Fig 2B. (B) Relative enrichment of total and each of mRNA isoforms compared to eef‐1A.1 mRNA by RT–qPCR. Error bars indicate standard error of mean (n = 3).
-
CExamples of normalized Nanopore currents for unmodified and m6A‐modified in‐vitro transcribed sams‐3/sams‐4 RNAs at nucleotide positions −4 through +4 relevant to the m6A site. One hundred reads are plotted for each. Color codes are indicated.
-
DDistribution of mean (top) and standard deviation (bottom) of normalized Nanopore currents at nucleotide positions −4 through +4 relevant to the m6A site for 5,000 of the unmodified and 2,680 of the m6A‐modified sams‐3/sams‐4 RNAs and 85 of an endogenous sams‐3 mRNA isoform E2/E3L. Color codes are indicated. Central bands represent the median, boxes represent the 25th and 75th percentiles, and whiskers represent the lowest and highest values. A red line at position 0 in the top panel indicates a cutoff line (mean current = 1.7557) that discriminates between the unmodified and m6A‐modified sams‐3/sams‐4 RNAs with accuracies of 64.58% for both.
-
EMachine learning‐based classification of sams RNA reads in Nanopore direct RNA sequencing data. The statistics in (D) as well as duration time of the Nanopore current at nucleotide positions −50 through +50 of up to 80% of mapped reads from the unmodified and methylated in␣vitro transcribed sams‐3/sams‐4 and sams‐5 RNAs were pooled and used for training with a machine learning algorithm Gradient Boosting and accuracy of the classifier was tested with 20% of the mapped reads. The classifier was also applied to reads identified as unproductive sams‐3, sams‐4, or sams‐5 mRNA isoforms in the analysis of endogenous mRNAs shown in Fig 1. Numbers of reads used in the tests are indicated at the top; percentages of reads classified as “m6A‐modified” are shown with color code.
-
FRelative importance of the statistics at each position for Gradient Boosting.
