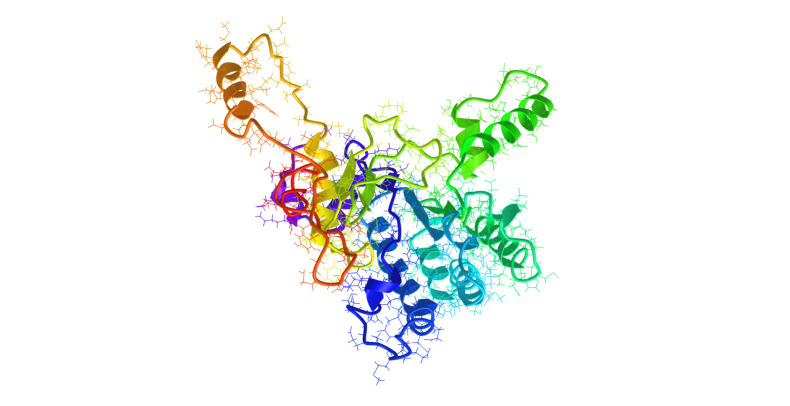
Figure 1. Example of a predicted structure used for annotating genes of unknown function.
DDB_G0275279 annotations from I-TASSER. Image of top seeded protein model 1 with TM-score = 0.76±0.10. This model is structurally closest to PDB: 3wqm, a Mycobacterium tuberculosis hydrolase / hydrolase inhibitor, TM-score 0.888 and a coverage (Cov) of 0.901, which is the number of structurally aligned residues divided by the total protein length. The best Enzyme Commission hit is to PDB:2VG1, a M. tuberculosis transferase with TM-score 0.603, two mapped active residues, and EC number 2.5.1.68. Gene Ontology annotations include GO:0008834, di-trans,poly-cis-decaprenylcistransferase activity, GO-score 0.74; GO:0008360, regulation of cell shape, GO-score 0.74; GO:0051301, cell division, GO-score 0.74; GO:0009252, peptidoglycan biosynthetic process, GO-score 0.74; GO:0071555, cell wall organization, GO-score 0.74; and GO:0016094, polyprenol biosynthetic process, GO-score 0.55.