TABLE 1.
Characterisation of peptides.
| Sequence | Code | Rt a | ESI-MSb | Kd (µM) ± sdc/EC50 (μM)d | ||
|---|---|---|---|---|---|---|
| Mcal. | Mmeas. | ERK2 | p38 | |||
| H-SLQRKKPPWLKLDIPSC-NH 2 | 1 | 2007.1 | 2006.4 | 1.8 ± 0.3 | 1.1 ± 0.3 | |
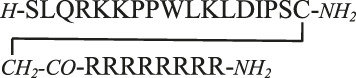
|
2 | 10.7 | 3315.0 | 3315.4 | 0.6 ± 0.3 | n.d. |
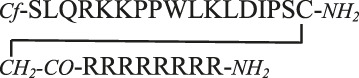
|
3 | 14.2 | 3673.4 | 3673.9 | n.d. | n.d. |
| H-KKPPWLKLDI-NH 2 | 4 | 12.2 | 1236.5 | 1236.3 | 15.0 ± 1.1 | n.d. |
| H-RRPPWLRLDI-NH 2 | 5 | 12.8 | 1319.8 | 1319.9 | 19.5 ± 2.6 | 10.0 ± 1.1 |
| H-RRPPWLRLDIRR-NH 2 | 6 | 11.4 | 1633.0 | 1632.6 | 5.2 ± 1.4/12.9d | n.d./3.7d |
| Cf-RRPPWLRLDIRR-NH 2 | 7 | 12.2 | 1991.3 | 1991.2 | n.d. | n.d. |
| H-RRRPPWLRLDIRR-NH 2 | 8 | 10.8 | 1788.1 | 1788.1 | 1.3 ± 0.1/3.5d | 1.4 ± 0.3/1.3d |
| Cf- RRRPPWLRLDIRR-NH 2 | 9 | 13.3 | 2146.4 | 2146.1 | n.d. | n.d. |
| Dabcyl-RRRPPWLRLDIRRK-NH 2 | 10 | 13.3 | 2167.5 | 2167.3 | 4.6 ± 0.6/8.3d | 1.0 ± 0.4/3.1d |
| Dabcyl-RRRPPWLRLDIRRK(Cf)-NH 2 | 11 | 14.7 | 2525.8 | 2526.4 | n.d. | n.d. |
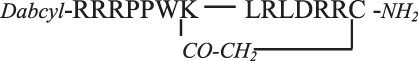
|
12 | 13.8 | 2197.2 | 2197.2 | 4.1 ± 0.3 | 1.7 ± 0.4 |
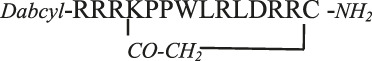
|
13 | 13.8 | 2197.2 | 2198.0 | 1.0 ± 0.2 | 1.0 ± 0.7 |
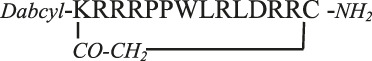
|
14 | 13.8 | 2197.2 | 2198.0 | 5.1 ± 0.4 | 2.1 ± 0.6 |
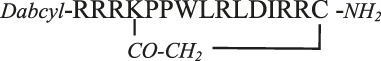
|
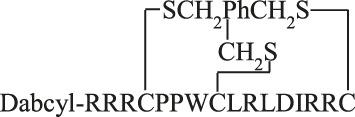
15 | 14.2 | 2310.3 | 2310.4 | 1.5 ± 0.3 | 1.2 ± 0.6 |
|
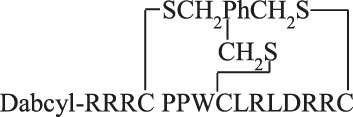
16 | 14.7 | 2462.3 | 2462.5 | 0.9 ± 0.2 | 1.4 ± 0.4 |
|
17 | 14.8 | 2349.2 | 2349.2 | 4.5 ± 0.6 | 3.2 ± 0.7 |
aAnalytical RP-HPLC was done on Nucleosil 120-3 C18 column (4.6 mm × 150 mm, 5 μm, 100 Å). The applied linear gradient elution was 0 min 0% B, 2 min 0% B, 22 min 90% B at 1 mL/min flow rate. The detection was carried on at λ = 220 nm.
bESI-MS.
cKd was determined by fluorescence polarization assay.
dEC50 values were determined by an in vitro MAPK activity assay, n.d. not determined.
All experiments were carried out at least in 3 independent measurements (Rt, retention time; Cf, carboxafluresceine).
