FIG 2.
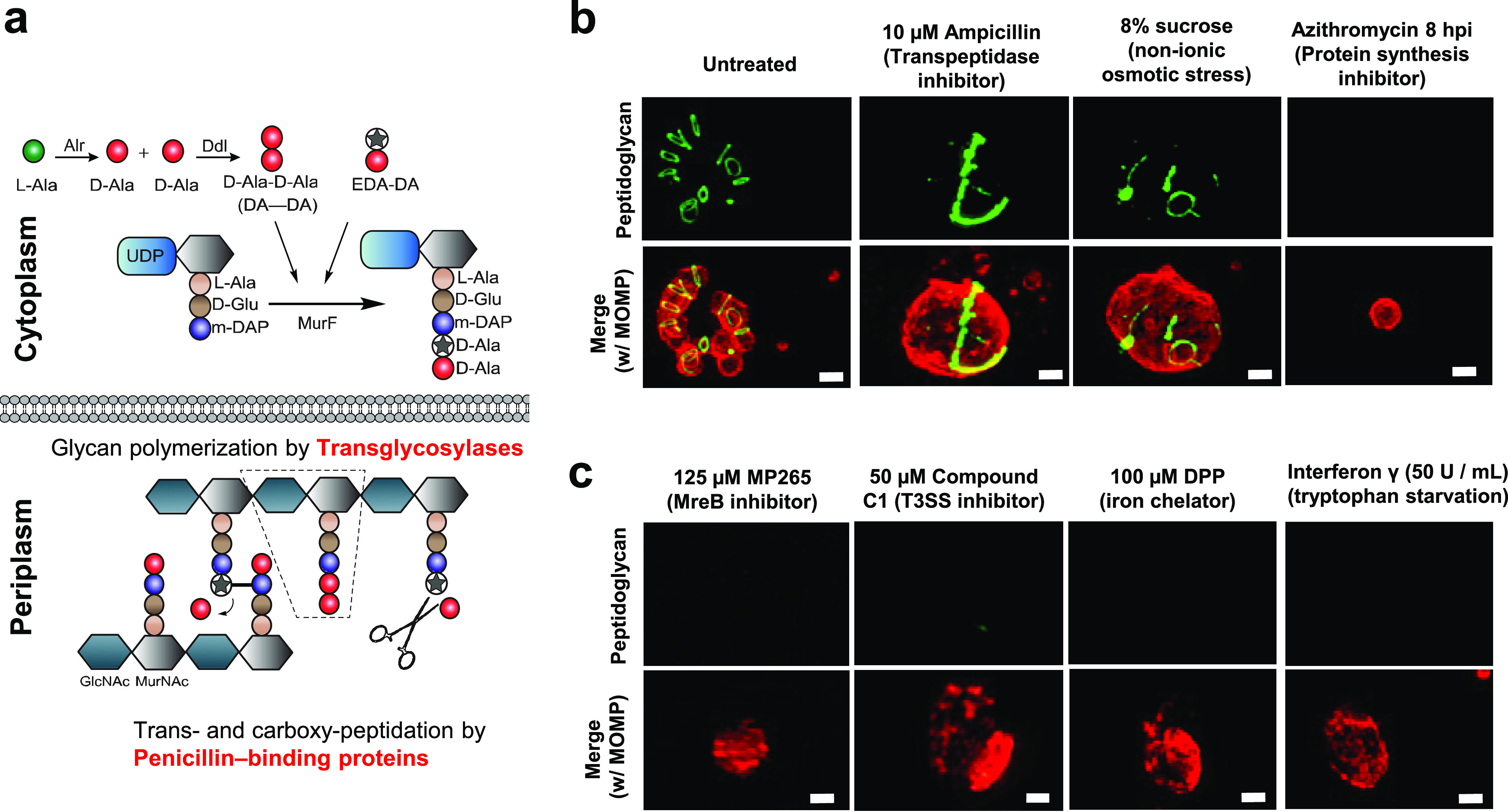
Aberrant Chlamydia forms differ in their abilities to synthesize peptidoglycan. (a) The Park nucleotide biosynthesis pathway in Gram-negative bacteria. Peptidoglycan can be labeled utilizing modified ethynyl-D-alanyl-D-alanine (EDA-DA) (green), which is subsequently incorporated into newly synthesized stem peptides (7). Alr, alanine racemase; Ddl, d-alanine-d-alanine ligase; MurE, UDP-N-acetylmuramoyl-l-alanyl-d-glutamate-diaminopimelate ligase; MurF, UDP-N-acetylmuramoyl-tripeptide-d-alanyl-d-alanine ligase. (b and c) Chlamydia-infected cells were grown under a variety of aberrance-inducing conditions. Peptidoglycan was labeled via click chemistry, and Chlamydia RBs and ABs were visualized utilizing a monoclonal antibody to the major outer membrane protein (MOMP) (red). Images are representative of 20 to 30 viewing planes over three independent experiments. ***, P < 0.001; ns, not significant. Bars, ∼1 μm.
