Figure 3.
Identification of Olig2-AS molecular signature by differential analysis
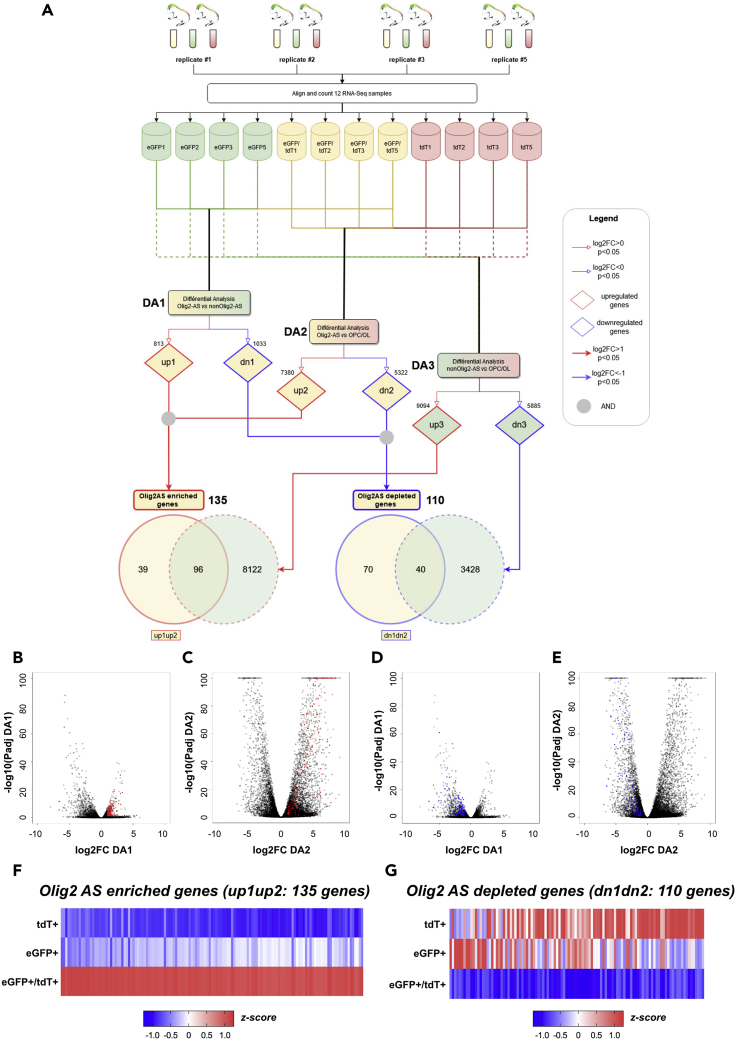
(A–G) (A) Flowchart representing the treatment and two-step comparative analyses performed on the RNA-seq data. The top half of the chart shows the 4 cell replicates of Olig2AS (yellow), nonOlig2AS (green), and OPC/OL (red), each obtained from the dissociation of two spinal cords isolated at P7. The bottom half represents the two consecutive comparisons performed between the three glial cell subtypes, and the numbers of genes found up-regulated (up, red) or down-regulated (dn, blue) in each pairwise analysis are indicated for each comparison. Numbers of genes found up-regulated (up1up2) or down-regulated (dn1dn2) in both DA1 and DA2 are indicated in solid line red and blue circles, respectively. Dashed line circles represent genes found up-regulated (red) or down-regulated (blue) in the DA3 comparison. Note that 39 genes were found up-regulated and 70 were found down-regulated specifically in Olig2AS compared with the two other glial cell populations. (B–E) Volcano plots of differentially expressed genes representing the log2FC and –log10padj from the differential analyses DA1 (B, C) and DA2 (D, E). Red dots in (B and C) represent enriched genes (up1up2; padj<0.05 and a log2FC > 1). Blue dots in (D and E) represent down-regulated genes (dn1dn2; padj<0.05 and a log2FC < 1). (F and G) Heatmap analysis representing the 135 genes up-regulated (F) and the 110 genes down-regulated (G) in Olig2-AS. All heatmaps are presented as Z score.
See also Figures S4, S5, and S6.