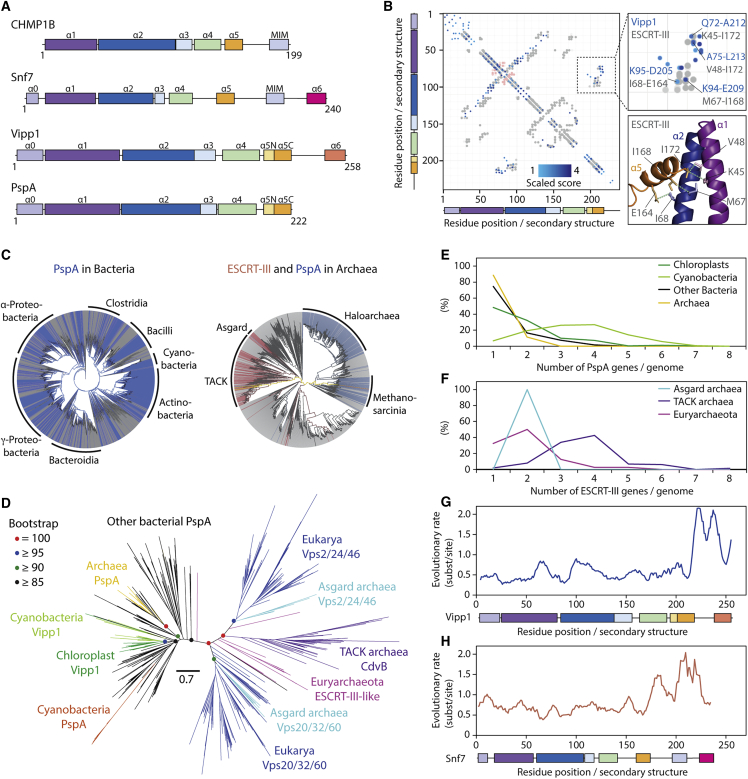
Figure 1.
ESCRT-III/Snf7/CdvB and PspA/Vipp1 proteins are part of the same superfamily
(A) ESCRT-III (human CHMP1B and yeast Snf7) and PspA (N. punctiforme Vipp1 and E. coli PspA) protein families are homologs and have a similar secondary structural organization (alpha helices are labeled α0–α6). Helices α6 have different colors because it is not clear if they are homologous across the PspA/Vipp1 and ESCRT-III/Snf7 families.
(B) A co-evolutionary analysis reveals similarities in the tertiary structure of PspA and ESCRT-III proteins. Left: plot shows a co-evolutionary residue contact map (using numbering based on the N. punctiforme Vipp1 sequence) super-imposed on ESCRT-III residue-residue distance data extracted from the experimentally determined structure of human CHMP3 (PDB: 3FRT). Extent of blue shading represents the strength in covariance between Vipp1 co-evolving residue pairs. Grey and red circles indicate intramolecular and intermolecular contacts (<5 Å) in the CHMP3 X-ray crystal structure, respectively. Top right: inset focuses on selected Vipp1 evolutionary coupled residues in blue clustering with known contacts derived from CHMP3 in gray. Bottom right: these contacts are mapped onto the CHMP3 structure.
(C) A phylogenetic analysis reveals a broad distribution of PspA/Vipp1 (blue) and ESCRT-III (red) homologs across bacteria (left, over 27,000 genomes) and archaea (right, over 1,500 genomes). Only a very few genomes encode both proteins (yellow). Genomes lacking both PspA/Vipp1 and ESCRT-III are presented in gray.
(D) Tree of the PspA/ESCRT-III superfamily colored according to phylogenetic groups inferred under the best-fitting LG+C30+G+F substitution model. A long branch separates the PspA/Vipp1 (left) and ESCRT-III (right) subfamilies. Scale bar represents expected substitutions per site.
(E and F) Number of copies of PspA/Vipp1 (top) and ESCRT-III/Snf7/CdvB (bottom) genes found per genome in different taxonomic groups. The analysis of the number of PspA in bacteria excludes cyanobacterial genes.
(G and H) Site-specific evolutionary rates in units of expected number of substitutions per site across PspA/Vipp1 and ESCRT-III/Snf7 protein families using N. punctiforme Vipp1 (top) and yeast Snf7 (bottom) as reference sequences.