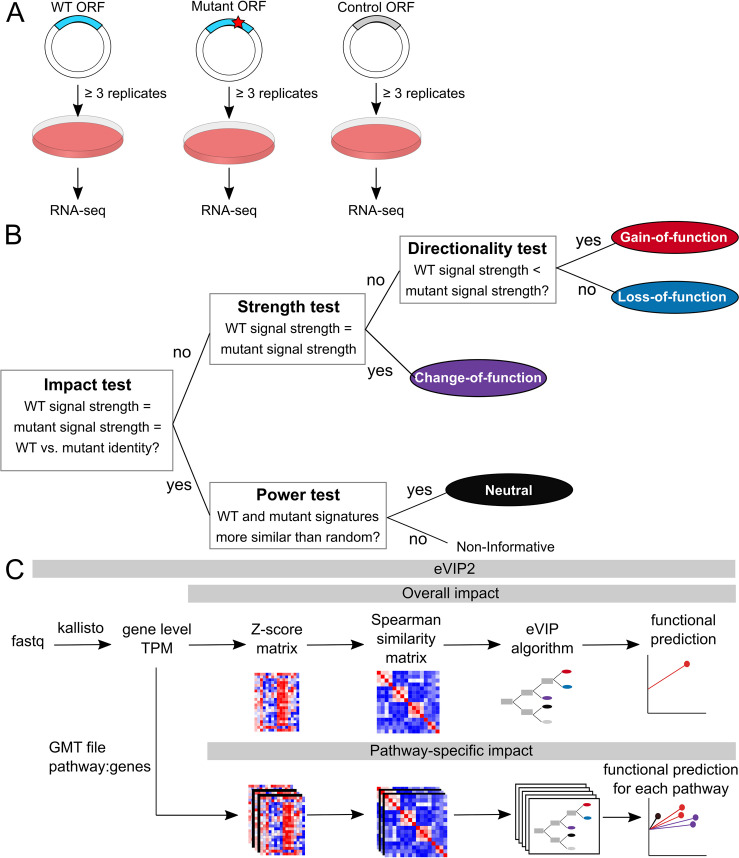
Fig 1. The eVIP algorithm uses RNA-seq data to predict the function of somatic mutations.
(A) Overview of the experimental approach (B) Schematic of the eVIP decision tree-based eVIP algorithm. The impact test is a Kruskal-Wallis test of the three distributions: wild-type replicate self-correlation, mutant replicate self-correlation, and wild-type versus mutant correlation. It outputs a Bonferonni-adjusted p-value, which represents the likelihood of mutation impact. Impactful mutations are then tested for their directional impact of gain-of-function (GOF) or loss-of-function (LOF). For non-impactful mutations, a “power test” determines whether the two signatures are similar to one another due to a real signal or due to noise. If they are similar, the mutation is considered to have a neutral impact [7]. (C) Overview of the eVIP2 pipeline which incorporates overall impact and pathway impact. The eVIP2 pipeline uses gene level counts (TPM) to predict the functional impact of a mutation. Many mutations can be processed in parallel. There is the option for eVIP Pathways, which predicts the impact of each mutation on a pathway.