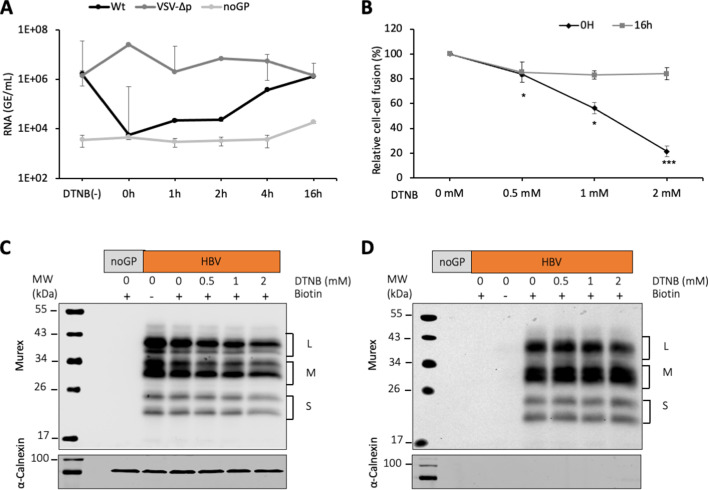
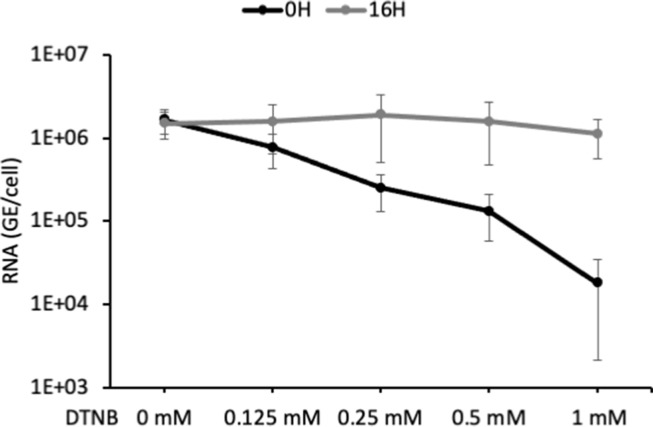
Figure 3. DTNB, a thiol-specific oxidizing reagent, inhibits HBV membrane fusion.
(A) 5,5-Dithiobis(2-nitrobenzoic acid) (DTNB) (2 mM) was added to the cell supernatant containing hepatitis delta virus (HDV) particles at the onset of infection (0 hr) or at the indicated times post-infection and was removed 8 hr later. VSV-∆p, that is, HDV particles generated with vesicular stomatitis virus-G glycoprotein (VSV-G GP) rather than hepatitis B virus (HBV), were used as the control for a virus entry process that is not affected by DNTB. As a negative control, pSVLD3 was co-transfected with an empty plasmid (referred to as ‘noGP’). At 7 days post-infection, HDV RNAs were extracted from infected cells and quantified by quantitative reverse transcription PCR (RTqPCR). The results are expressed after normalization with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNAs as means ± SD (N = 3) per ml of cell lysates containing 106 cells. The results of infection in the absence of DTNB are shown (DTNB(-)). (B) Huh7 ‘donor’ cells co-expressing HBV GPs and a luciferase marker gene driven by the HIV-1 promoter were co-cultured with Huh7-NTCP-tat ‘indicator’ cells that express HIV Tat protein. Different concentrations of DTNB were added at 0 hr vs at 16 hr after initiating the cell co-culture, as indicated. No cytotoxicity could be detected in these conditions (Figure 1—figure supplement 2). The luciferase activity induced by fusion between donor and indicator cells was then measured 24 hr later. Fusion mediated by HBV GPs without DTNB was taken as 100%. The graphs represent the average of four independent experiments. (C, D) Huh7 cells transfected with pUC19 (noGP) or the pT7HB2.7 (HBV) plasmids were incubated with dimethyl sulfoxide (DMSO) (0) or increasing doses of DTNB (0.5, 1, and 2 mM) for 16 hr prior to incubation with biotin for 30 min at 4°C. Biotin was omitted from one sample (-) and served as a negative control for non-specific binding of proteins to streptavidin. Cells were subsequently lysed and the biotinylated surface proteins were captured by streptavidin agarose. Total (C) and biotin-labeled proteins (D) were then analyzed by western blot using anti-HBsAg (Murex) and anti-calnexin antibodies. Calnexin detection was used as a control for the cytoplasmic protein marker, showing the integrity of the cell membrane, as shown in these representative western blots. The molecular weight markers (kDa) are shown on the left.