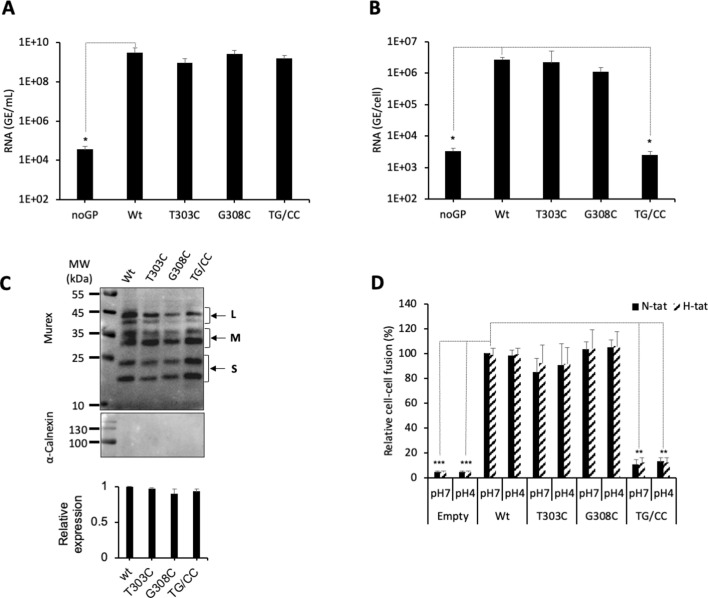
Figure 5. Evidence for a functional role of the CSD in the region 294–317 of the HBV S GP.
(A) Huh7 cells were co-transfected with pSVLD3 plasmid coding for hepatitis delta virus (HDV) RNPs and plasmids coding for wild-type (wt), single, or double mutant (TG/CC) hepatitis B virus glycoproteins (HBV GPs). As control, pSVLD3 was co-transfected with an empty plasmid (referred to as ‘noGP’). At day 9 post-transfection, the cell supernatants were harvested, filtered, and the extracellular RNA extracted and purified before quantifying HDV RNAs by quantitative reverse transcription PCR (RTqPCR). HDV RNA levels in GE (genome equivalent) are expressed as means ± SD (N = 4) per ml of cell supernatants. (B) HDV particles were used to infect Huh7-NTCP cells, which were grown for 7 days before total intracellular RNA was purified. The results of HDV RNA quantification by RTqPCR are expressed after normalization with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNAs as means ± SD (N = 4) per ml of cell lysates containing 106 cells. (C) Detection of GP mutants at the cell surface by biotinylation. Huh7 cells expressing wt or mutant HBV GPs were biotinylated for 30 min at 4°C and then processed biochemically. Cell lysates were subjected to streptavidin pull-down prior to western blot analysis using anti-HBsAg antibody (Murex). The molecular weight markers (kDa) are shown on the left. Calnexin detection was used as a control for the cytoplasm protein marker, showing the integrity of cell membrane, as shown in this representative western blot. The relative quantification of cell-surface GP expression compared to wt quantified from western blot analyses (means ± SD; N = 3) is shown below. See the quantification of total HBV GP expression in Figure 1—figure supplement 4. (D) Huh7 ‘donor’ cells co-expressing wt or mutant HBV GPs and a luciferase marker gene driven by the HIV-1 promoter were co-cultured with either Huh7-tat (H-tat) or Huh7-NTCP-tat (N-tat) ‘indicator’ cells that express HIV Tat protein. After 24 hr, the cells were treated at pH 4 or pH 7 for 3 min. The luciferase activity induced by the fusion between the donor and indicator cells was measured 24 hr later. Fusion mediated by wt GP at pH 7 with Huh7-NTCP-tat cells was taken as 100%. The bars represent the means (N = 4). Error bars correspond to standard deviations.