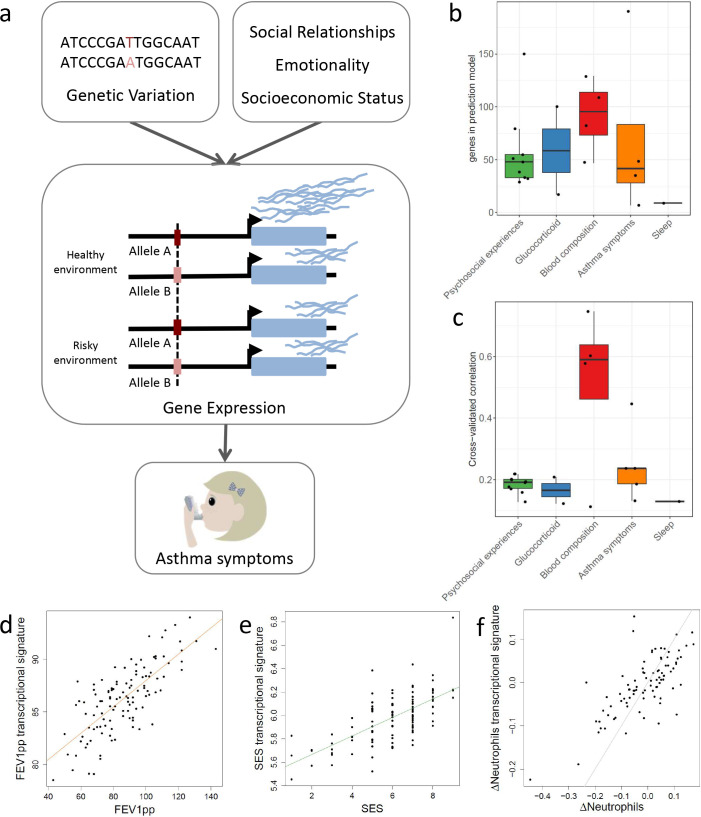
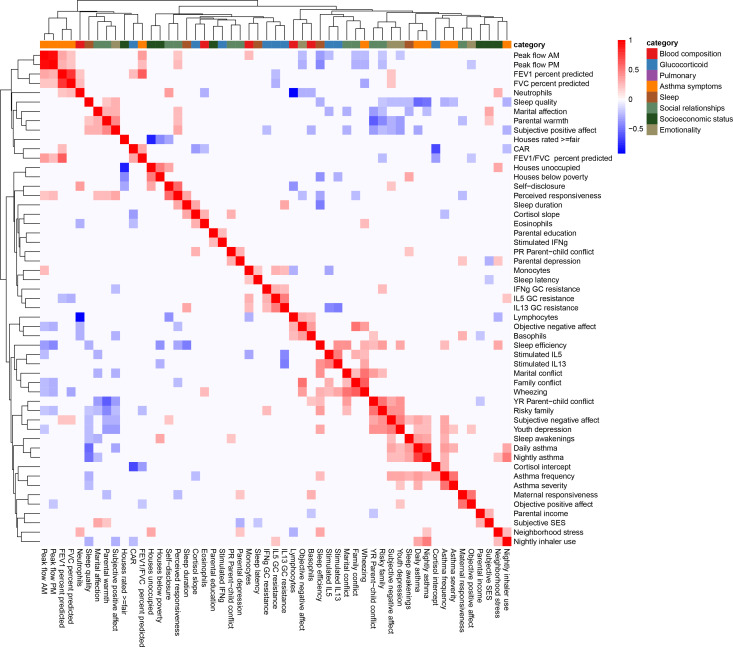
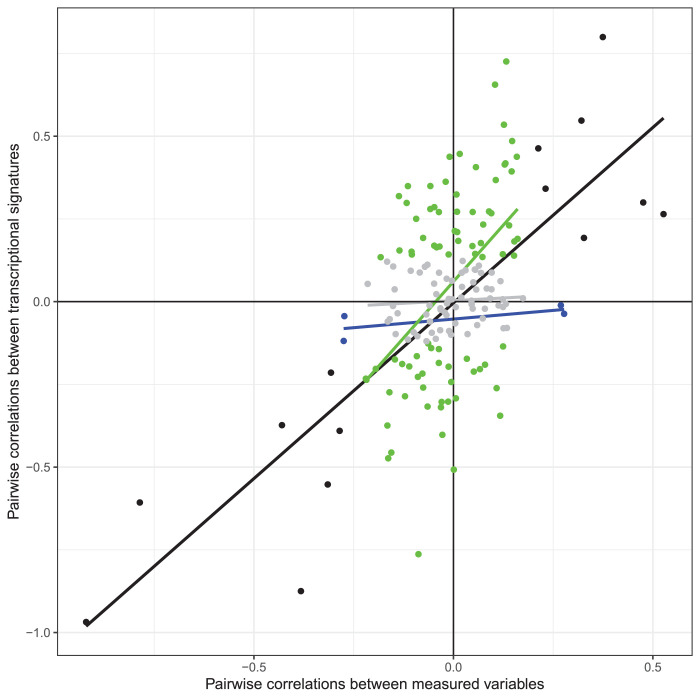
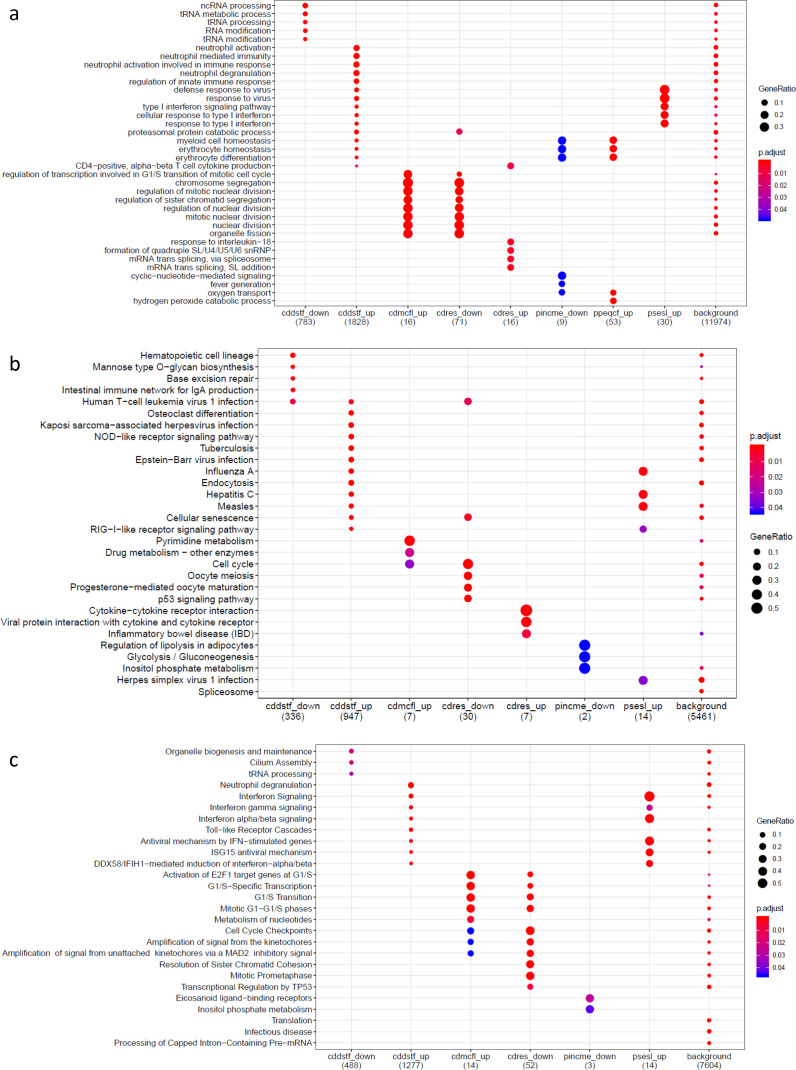
Figure 1. Transcriptional signatures of psychosocial experiences and asthma symptoms.
(a) Central hypothesis. (b) Number of genes in elastic net regression models that explain at least 1% of variance. Colors represent different categories of variables. (c) Pearson’s correlations between cross-validated transcriptional signatures and measured variables for elastic net regression models that explain at least 1% of variance. (d) Forced expiratory volume in one second [FEV1] percent predicted transcriptional signature model fit (Pearson’s rho = 0.76, p<0.001). (e) MacArthur socioeconomic status transcriptional signature model fit (Pearson’s rho = 0.67, p<0.001). (f) Longitudinal change in observed neutrophils (x axis) and longitudinal change in transcriptional signature of neutrophils (y axis) (Pearson’s rho = 0.72, p<0.001, gray = identity line).