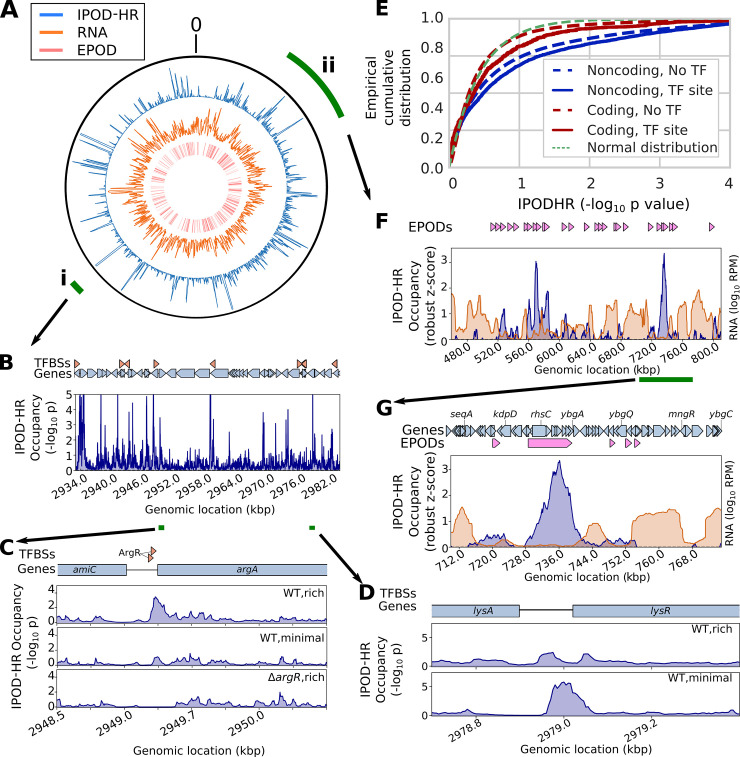
Fig 2. IPOD-HR profiles reveal rich high-resolution occupancy dynamics and large-scale structural features across the chromosome.
(A) Outer track: IPOD-HR occupancy (robust Z-scores, 5-kb moving average); middle track: total RNA read density (5-kb moving average); inner track: locations of inferred EPODs. The outer green wedges mark the portion of the chromosome shown in subsequent panels. The origin of the coordinate system is oriented at the top of the plot. All data in this figure are for the “WT,rich” condition unless otherwise noted. (B) IPOD-HR occupancy measured during growth in glucose RDM, in the vicinity of wedge i from panel A. Green segments below the genomic coordinates indicate the regions highlighted in panels C–D. (C) Condition-dependent occupancy changes at the ArgR binding sites upstream of argA. (D) Identification of condition-specific occupancy of a likely LysR binding site between lysA and lysR. (E) Cumulative histograms showing RNA polymerase ChIP-subtracted IPOD-HR occupancy in coding vs. noncoding regions and at sites that match known TFBSs from RegulonDB [7], compared with the curve that would be expected from a standard normal distribution of scores. Additional descriptive statistics and significance calls are given in S1 Table. (F) Occupancy (blue) and total RNA abundance (orange) for a selected sector of the genome (wedge ii from panel A), showing the presence of several EPODs in regions corresponding to low RNA abundance; rolling medians over a 5-kb window are plotted, with RNA read densities shown in units of RPM. (G) Magnification of the region highlighted by the green bar in panel F, illustrating a silenced region in and around rhsC, alongside flanking areas of low IPOD-HR occupancy and high transcription. A 5-kb rolling median is plotted. ChIP, chromatin immunoprecipitation; EPOD, extended protein occupancy domain; IPOD-HR, in vivo protein occupancy display—high resolution; RDM, rich defined medium; RPM, reads per million; TF, transcription factor; TFBS, transcription factor binding site; WT, wild-type.