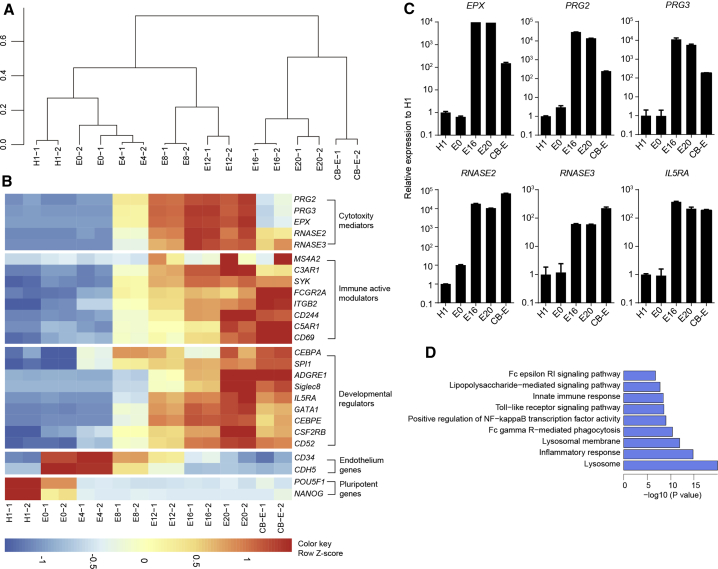
Figure 2.
Transcriptional analysis of H1-derived eosinophils
(A) Dendritic hierarchical clustering of global gene expression of H1 cells and induced cells harvested on E0, E4, E8, E12, E16, and E20. Cord-blood-derived naive eosinophils (CB-E) were used as a positive control. Notably, the cells on E0 were sorted for CD34+ cell population for transcriptional analysis, while the whole cell culture was collected for transcriptional analysis on E4, E8, E12, E16, and E20. For cells at each time point, duplicate experiments were performed.
(B) Heatmaps of indicated gene expression, including cytotoxicity mediators, immune active modulators, developmental regulators, endothelium genes, and pluripotency genes in H1, CB-E, and induced cells harvested on E0, E4, E8, E12, E16, E20, and CB-E based on Z score from FPKM.
(C) Real-time qPCR analysis of key eosinophil gene expression in cells recovered on D0, E0, E16, and E20. Cord-blood-derived naive eosinophils (CB-E) were used as a positive control (n = 2). Data are shown as mean value ± SD. Similar results were obtained in three independent experiments.
(D) GO term and KEGG analysis showing GO terms and KEGG pathways that were upregulated in H1-derived eosinophils collected on E20 compared with H1.