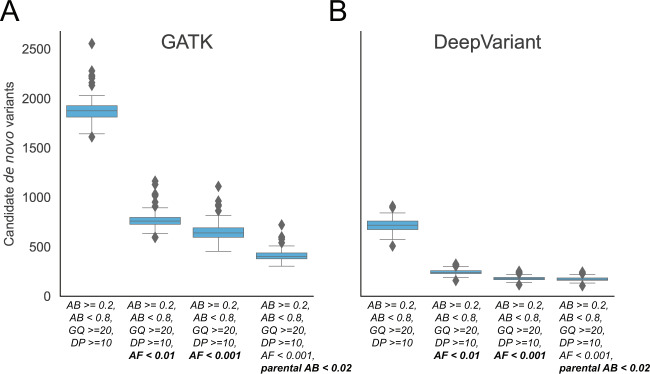
Fig. 4. Candidate autosomal de novo variants per genome identified by GATK and DeepVariant outside of low-complexity regions.
A cohort of 94 WGS trios from the Rare Genomes Project were screened for candidate de novo mutations using GATK (A) and DeepVariant (B). Variants lying in low-complexity regions were excluded. The leftmost boxplot within each subplot requires a depth ≥10, an allele balance between 0.2 and 0.8 along with a genotype quality (GQ) ≥20. Lines within the boxplot are determined from the quartiles of the data. The next box requires that the allele frequency in gnomAD is <0.01. The third box lowers the allele-frequency cutoff in gnomAD of <0.001. The final box excludes candidate de novo variants where the allele balance (of the homozygous call) in the parent is ≥2%. Supplementary Fig. 13 presents the analogous plots when including low-complexity regions.