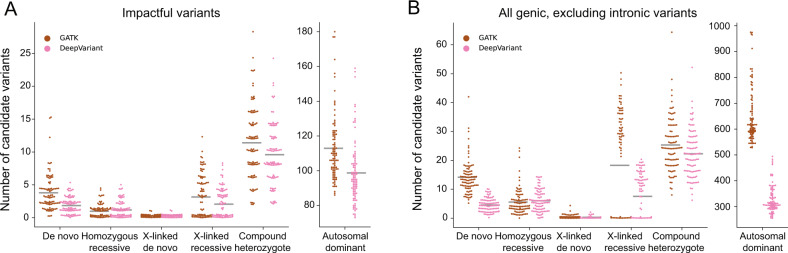
Fig. 5. The number of candidate variants that follow different inheritance modes per genome using two different variant callers.
A Only “impactful” variants as determined by slivar using annotations from VEP, snpEff, or bcftools are shown. B The set of variants is extended to include synonymous, UTR, and conserved intron regions (but not all intronic). Counts for autosomal dominant variants are shown in a separate plot due to the much larger numbers. Each dot represents the number of candidate variants (y-axis) passing the inheritance mode (x-axis), genotype-quality, population allele-frequency, and allele-balance filters for a single family. Gray bars indicate the mean number for each class and inheritance mode. We show Fig. 5A for the sarcoma replication cohort in Supplementary Fig. 14.