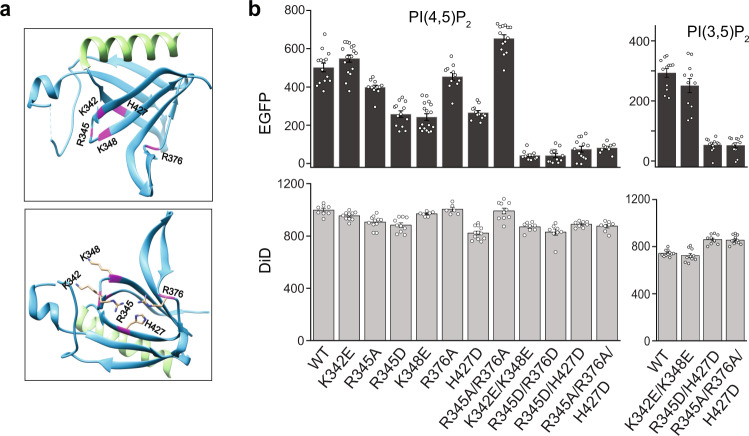
Fig. 4. Mutations in the PH domain differentially abolish ARHGEF3 binding of PI(4,5)P2 and PI(3,5)P2.
a Ribbon representation of the structure of ARHGEF3 PH domain (PDB ID: 2Z0Q;22 image created with Pymol v2.5), with residues predicted to interact with PIP highlighted in magenta and the sidechains of those residues shown in the lower panel. b HEK293 cells were transiently transfected with EGFP-ARHGEF3 containing various mutations at the residues highlighted in (a), and the lysates were subjected to SiMPull assays against vesicles containing PI(4,5)P2 or PI(3,5)P2. Numbers of fluorescent molecules per image area are shown in the graphs. Data are presented as mean ± SEM. N ≥ 10 images for EGFP; N ≥ 6 images for DiD. Source data are provided as a Source Data file, which lists the exact N number for each data point. Each assay was performed with at least three independent transfections with similar results.