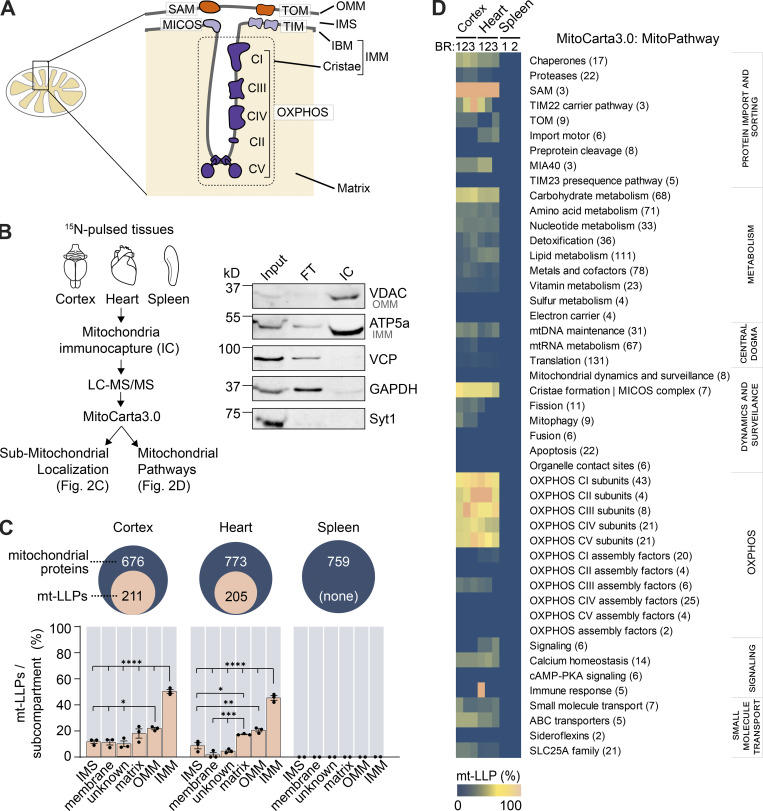
Figure 2.
LLPs are preferentially associated with the IMM. (A) Schematic illustrating mitochondrial subcompartmentalization and relevant protein complexes. (B) Workflow to immune-capture mitochondria. Cortex, heart, and spleen from 15N-pulsed mice were homogenized using a gentleMACS system, and dissociated mitochondria were purified by immuno-capture using anti-Tom20 antibodies (Miltenyi Biotec). Purified mitochondria were analyzed using LC-MS/MS, mitochondrial proteins were filtered using MitoCarta 3.0, and mt-LLPs were grouped and examined based on their submitochondrial localization and Mitochondrial Pathway assignments. FT, flow through; IC, immunocapture; IBM, inner boundary membrane. (C) The total number of mitochondrial proteins identified based on 15N and 14N MS/MS spectral matches per tissue (blue) along with the number of identified 14N mt-LLPs (tan). mt-LLPs are enriched at IMM in both heart and cortical extracts, but not in the other subcompartments. None of the mitochondrial proteins in spleen were identified as mt-LLPs. (D) MitoPathway analysis of mt-LLPs. mt-LLPs are enriched in several protein complexes including OXPHOS, Tim22, TOM, SAM, and the cristae formation (i.e., MICOS complex). BR, biological replicate. Data in C are mean ± SEM; for C and D, n = 3 mice for cortex and heart, 2 for spleen. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Kruskal–Wallis ANOVA with Tukey’s multiple comparisons test. See also Fig. S2 and Table S2.