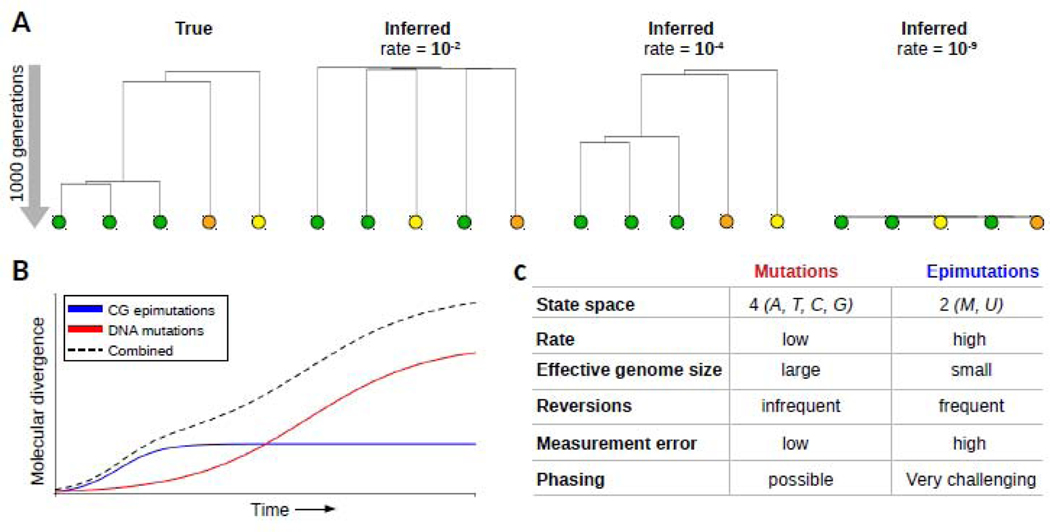
Fig. V: Epimutations as an evolutionary clock.
(A) The inclusion of epimutation data can improve the temporal resolution of phylogenetic analyses of the recent past To illustrate this we simulated an ultmetric tree (true, left) with five lineages and a depth of 1000 generations. Conditional on the tree topology, we simulated a donally reproducing species forward in time, and introduced epimutations at a rate of 10J, 10- and 10−5, per site, per haploid genome per generation. mCG divergence observed between the tips were used as input to construct a neighbor-joining tree for each scenario. On this time-scale, a high rate (10−2) or a low rate (10−9) lead to incorrect inferences of topology and relative branch lengths With a rate of 10−4, which is the approximate rate estimated in plants, the epimutation data contained sufficient information to reconstruct the true tree With calibrated epimutation rates, it will be possible to estimate branch ages and branch point times in recent phylogemes The color of the circles emphasize the original cluster membership of samples in the true tree (B) Schematic showing that CG epimutations diverge relatively quickly over time and saturate early By contrast, DNA mutations accumulate slowly and saturate late Both molecular markers capture different segments of evolutionary time. Future methods should integrate CG epimutations and DNA mutations to cover the full time-scales. (C) The incorperation of epimutation data into existing phylogenetic and population genetic models will be challenging as epimutations differ in key aspects from DNA mutations. These differences should inform the development of new models.