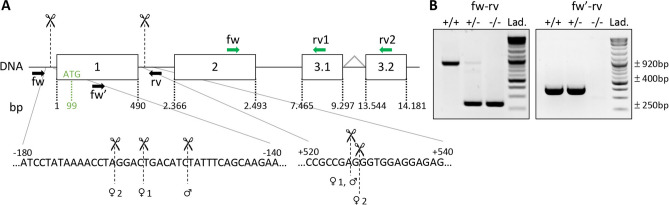
Figure 2.
CRISPR/Cas9-based editing of MarvelD3 gene. (A) Schematic representation of the MarvelD3 locus. The MarvelD3 gene comprises four exons (boxes), the two first are common while either exon 3.1 or 3.2 are alternatively spliced to generate the two isoforms (MD3.1 and MD3.2). Numbers below the boxes indicate the position of the exons limits with respect to the transcription initiation site (+ 1). Position of the ATG (99 bases downstream of the + 1) in the first exon is also indicated. Scissors and the vertical dashed lines indicate the approximate region where the cuts occurred. Precise localization of the cuts is provided below for the three individuals used to derive the colony. Localisation of genotyping primers are represented by black arrows (fw-rv and fw′-rv); primers to assess MarvelD3 expression by RT-qPCR are shown in green (fw-rv1 = MD3.1 and fw-rv2 = MD3.2). (B) Illustrative genotyping results for wild-type (+ / +), heterozygous (+ /−) and knockout (−/−) mice, obtained with two sets of primers. Primers fw-rv, located outside of the targeted region (exon 1), can generate PCR products of ± 920 bp (wild-type allele: +), and ± 250 bp (deleted allele: -). Primers fw′-rv will only amplify a band of ± 400 bp on the wild-type allele (+ / + and + /−). Lad., DNA ladder.