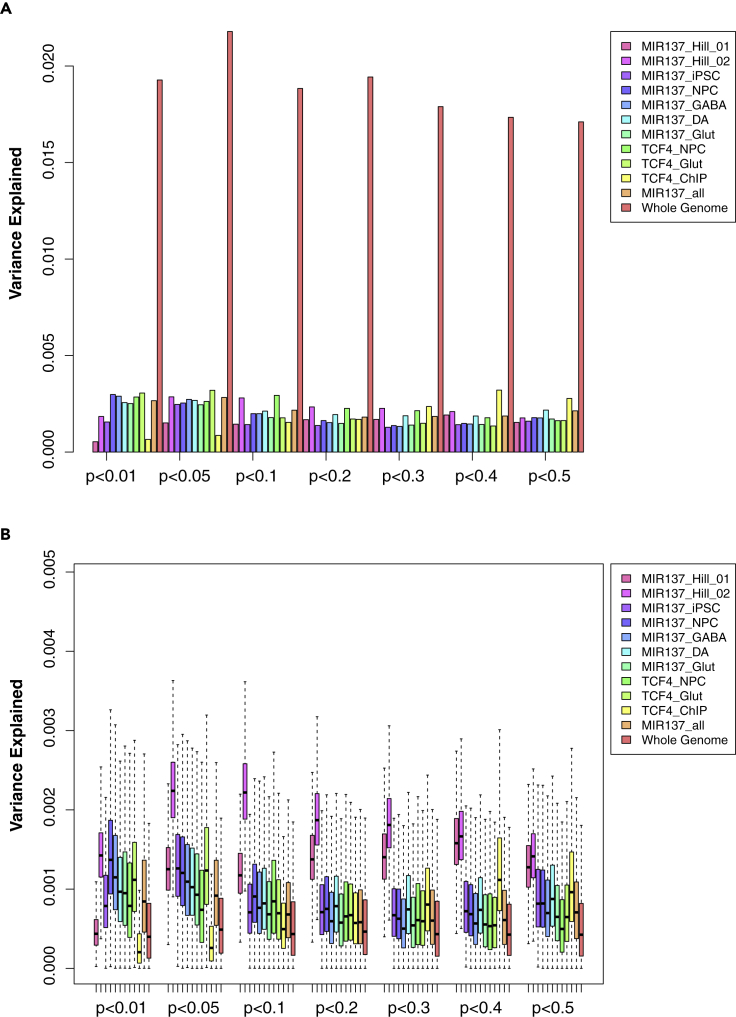
Figure 5.
PRS analysis of different SNP lists of MIR137 and TCF4 target genes annotated in a 20-kb window for the Han Chinese sample
(A) PRS result using LD-pruned SNPs (r2 < 0.2) for each gene list. The variance explained in the target sample is based on risk scores derived from an aggregated sum of weighted SNP risk allele effect sizes estimated from the discovery samples at seven significance thresholds (p < 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5). The y axis indicates the percentage of phenotypic variance explained by the PRS (Nagelkerke's pseudo R2).
(B) The permutation (N = 1,000) PRS results based on randomly selected 1,500 SNPs for each gene set. Data are presented as mean ± SEM. Gene sets in (A) and (B) were described in Table 1.