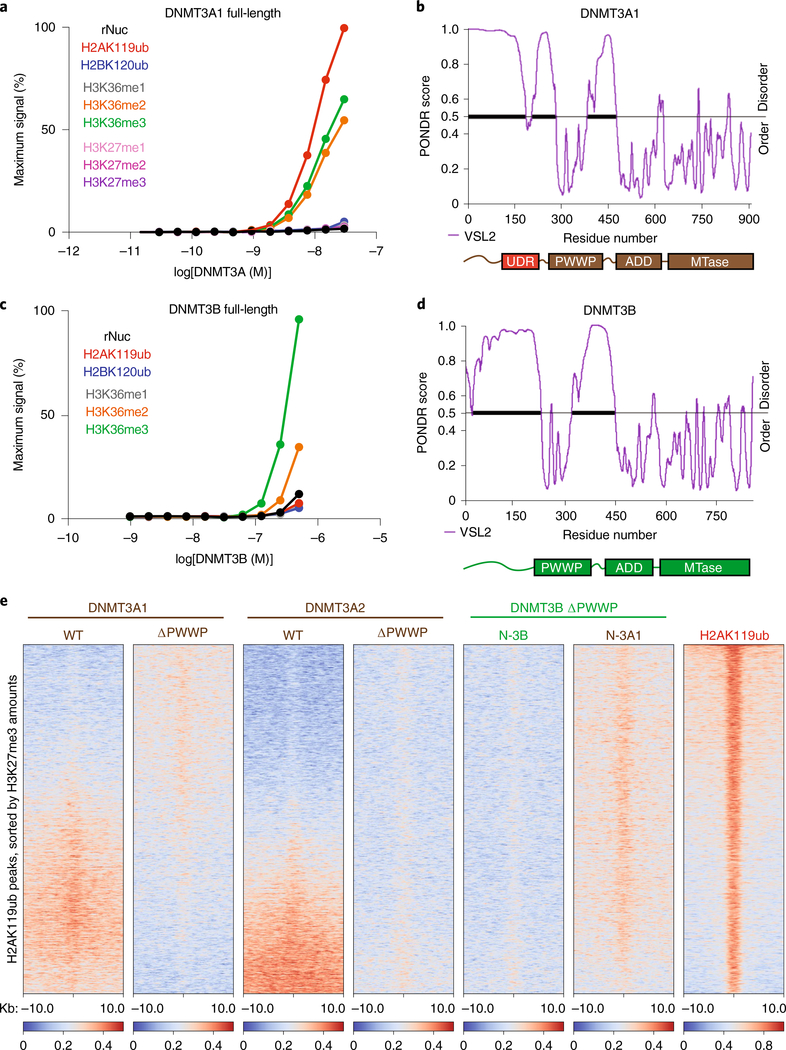
Fig. 3 |. DNMT3A interacts directly with H2AK119ub through an N-terminal UDR.
a, AlphaLISA counts for interaction of GST-tagged full-length DNMT3A1 titrated against modified (as indicated) or unmodified (rNuc) nucleosomes. Data are mean values from replicates and are representative of two independent experiments. b, Graph of intrinsic disorder for DNMT3A1. PONDR VSL2 scores are indicated on the y axis and the amino acid positions are indicated on the x axis, with the domain structure of DNMT3A1 shown below. c, AlphaLISA counts for interaction of GST-tagged full-length DNMT3B titrated against modified (as indicated) or unmodified (rNuc) nucleosomes. Data are mean values from replicates and are representative of two independent experiments. d, Graph of intrinsic disorder for DNMT3B. PONDR VSL2 scores are indicated on the y axis and the amino acid positions are indicated on the x axis, with the domain structure of DNMT3B shown below. e, Enrichment heat map depicting ChIP-seq normalized reads centered at H2AK119ub peaks ± 10 kb (n = 16,064), sorted by H3K27me3 amounts for DNMT3A1 wild type, DNMT3A1 ΔPWWP, DNMT3A2 wild type, DNMT3A2 ΔPWWP, DNMT3B ΔPWWP (N-3B) and chimeric DNMT3B ΔPWWP containing the amino-terminal residues 1–219 of DNMT3A1 (N-3A1), and H2AK119ub.