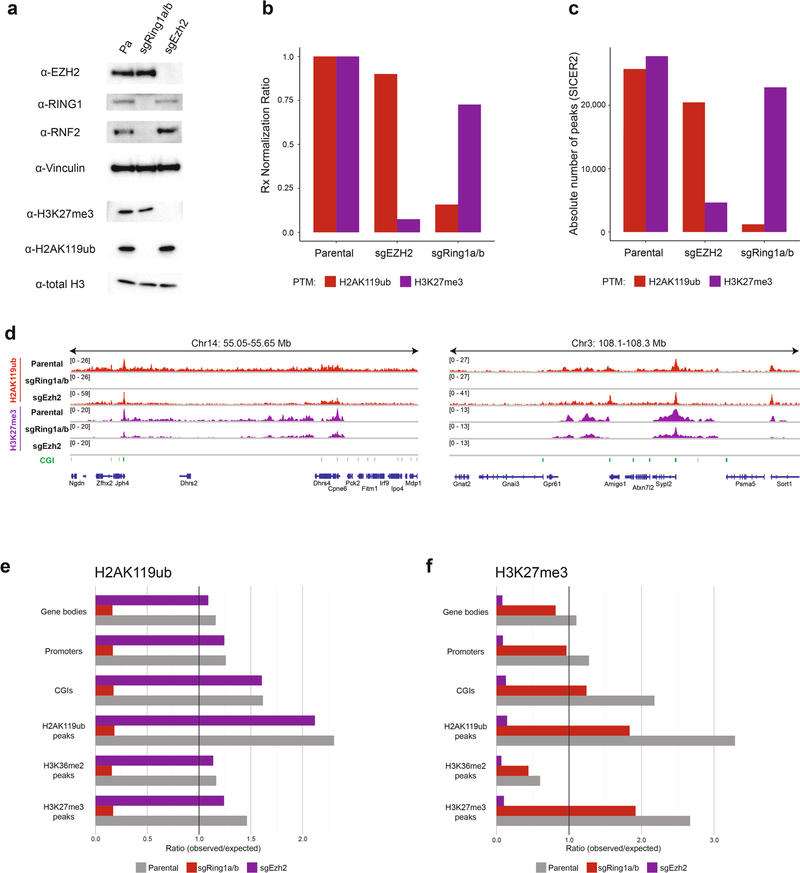
Extended Data Fig. 5 |. Perturbation of PRC1 and PRC2 in mouse MSCs.
a) Immunoblots of lysates generated from parental, sgRing1a/b, and sgEzh2 mouse MSCs. Vinculin and total H3 were used as loading controls. Data are representative of two independent experiments. b) Ratios of ChIP-seq reads for H2AK119ub and H3K27me3 between target chromatin (mouse) and reference spike-in chromatin (Drosophila) in parental, sgRing1a/b, and sgEzh2 mouse MSCs. c) Absolute number of ChIP-seq peaks for H2AK119ub and H3K27me3 called by SICER2 in parental (first replicate), sgRing1a/b, and sgEzh2 mouse mMSCs. d) Genome browser representation of ChIP-seq normalized reads for H2AK119ub and H3K27me3 in parental, sgRing1a/b, and sgEzh2 mouse MSCs at chromosome 14: 50.05–55.65 Mb and chromosome 3: 108.1–108.3 Mb. CGIs (green) and genes from the RefSeq database are annotated at the bottom. e) Rx-adjusted ratio of observed-to-expected ChIP-seq reads for H2AK119ub in annotated genomic regions in parental, sgRing1a/b, and sgEzh2 cells. Numbers of expected reads were generated assuming equivalent genomic distribution to input. f) Rx-adjusted ratio of observed-to-expected ChIP-seq reads for H3K27me3 in annotated genomic regions in parental, sgRing1a/b, and sgEzh2 cells. Numbers of expected reads were generated assuming equivalent genomic distribution to input.