Figure 5.
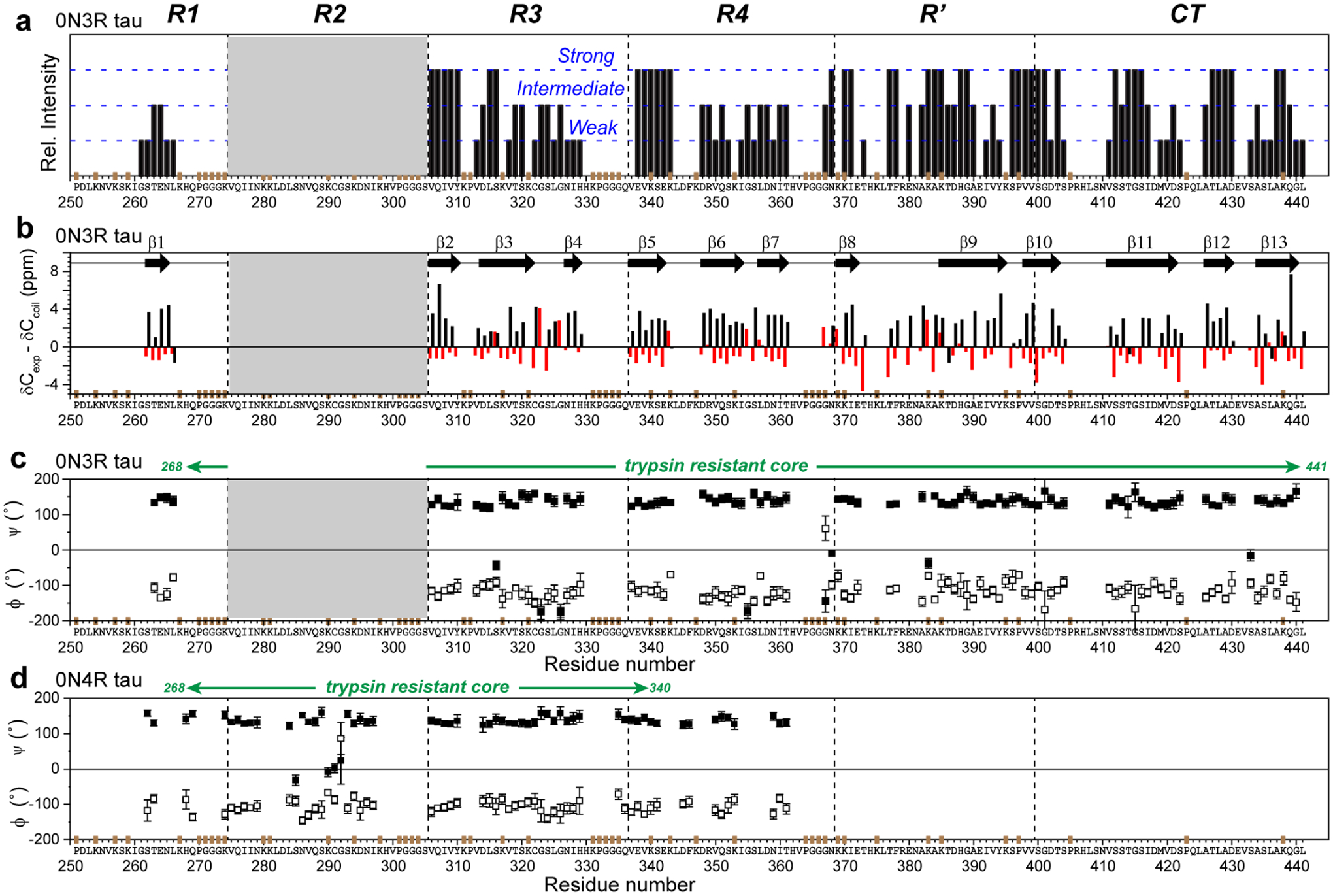
Backbone conformation of the 0N3R tau fibril core obtained from 13C and 15N chemical shifts. (a) Relative peak intensities of assigned residues, categorized qualitatively as strong, intermediate and weak based on comparing resolved peaks of the same type. Strong peaks indicate rigid and ordered residues. R1 residues have weaker intensities than residues in other domains. The positions of Pro (P), GGG triplets, and unlabeled Lys (K) residues are indicated in brown on the x-axis. (b) Cα and Cβ secondary chemical shifts. Negative Cα and positive Cβ secondary shifts are indicative of a β-strand conformation. Thirteen β-strands can be identified (arrows). Since not all residues are assigned, the exact number of distinct β-strands in the protein may differ. (c) Backbone torsion angles predicted from the measured chemical shifts27. (d) (ϕ, ψ) torsion angles of heparin-fibrillized 0N4R tau for comparison24.
