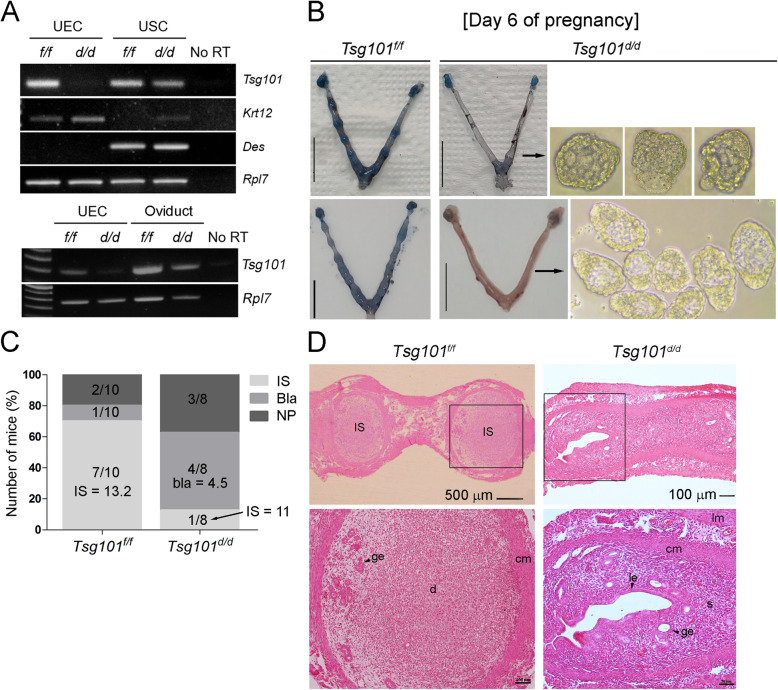
Fig. 1.
Compromised implantation in day 6 pregnant Ltf-iCre/Tsg101f/f mice. (A) Uterine epithelium-specific deletion of floxed Tsg101 gene by Ltf-iCre recombinase was confirmed by RT-PCR. Uterine epithelial cells (UECs) and uterine stromal cells (USCs) were isolated from 8 to 10-week-old random cycling Tsg101f/f or Ltf-iCre/Tsg101f/f (Tsg101d/d) mice. Deletion of Tsg101 in Tsg101d/d UECs was confirmed. Keratin 12 (Krt12) and desmin (Des) were used as marker genes for UECs and USCs, respectively. Ribosomal protein L7 (Rpl7) is a housekeeping gene and was used as an internal control. Five mice were used for cell isolation in each group. The bottom panel shows expression of Tsg101 in Tsg101f/f and Tsg101d/d oviducts. (B) Uteri of day 6 pregnant Tsg101f/f or Tsg101d/d mice. Mice (10–11-week-old) received 2.5 IU of eCG and hCG and were bred with stud male mice. On day 6 of pregnancy, the mice received a blue dye injection to demarcate the implantation sites (IS). Uteri from mice without IS were flushed. Scale bar, 1 cm. (C) Tsg101f/f (n = 10) or Tsg101d/d (n = 8) with variable pregnancy outcomes on day 6. Most of the Tsg101f/f mice had IS (average number = 13.3), whereas 50% of the Tsg101d/d mice showed unimplanted blastocysts upon uterine flushing. IS, mice with implantation sites; Bla, mice with unimplanted blastocysts; NP, not pregnant. Average number of implantation sites or blastocysts is shown on the graph. (D) Histological analysis of day 6 pregnant uterine sections. A representative set of figures is shown. Paraffin-embedded sections were stained with hematoxylin and eosin. IS, implantation site; ge, glandular epithelium; cm, circular muscle; lm, longitudinal muscle; le, luminal epithelium; s, stroma. Areas demarcated with a black square are magnified in the lower panels