Figure 1.
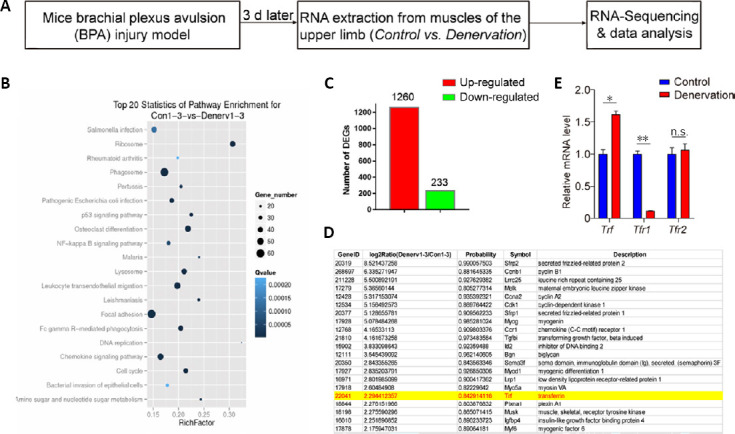
Identification of transferrin as one of the upregulated genes in denervated skeletal muscles.
(A) Schematic of the experimental design for the identification of denervated skeletal muscle-related genes in the brachial plexus avulsion injury mouse model and RNA deep-sequencing. (B) Kyoto Encyclopedia of Genes and Genomes pathway analysis and the top 20 enriched differential signaling pathways between control and denervated skeletal muscles. (C) Quantification of the total number of DEGs between control and denervated skeletal muscles. (D) Representative list of significantly upregulated genes, including transferrin and 20 other genes, in denervated skeletal muscles. (E) Quantitative reverse transcription-polymerase chain reaction analysis indicates significantly increased expression of Trf, but decreased expression of the Tfr1 gene, in denervated skeletal muscles. The expression of the Tfr2 gene was not changed. Data are expressed as the mean ± SEM (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). DEGs: Differentially-expressed genes; n.s.: not significant; Trf: transferrin; Tfr1: transferrin receptor 1; Tfr2: transferrin receptor 2.
