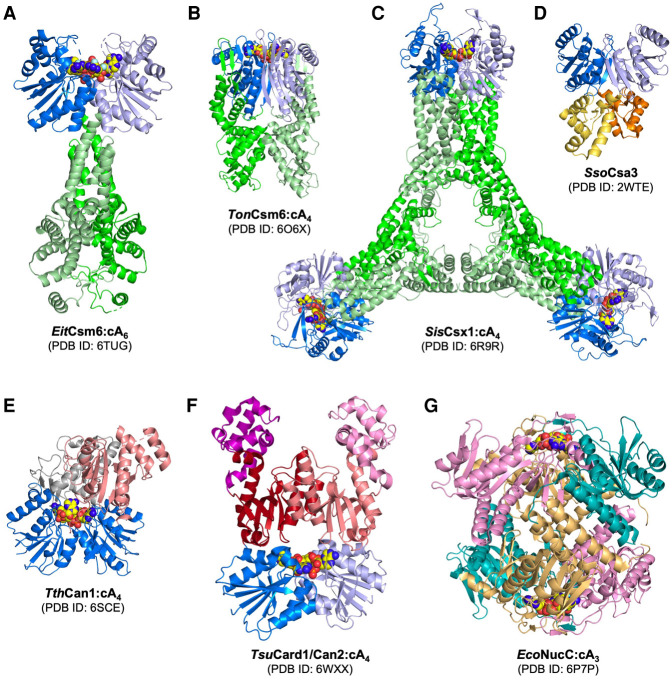
FIGURE 2.
Structures of type III CRISPR ancillary proteins. (A) E. italicus (Eit) Csm6 dimer in complex with its cyclic hexa-adenylate (cA6) activator (shown in sphere form). EitCsm6 is a nonspecific ribonuclease containing CARF (dark and light blue) and HEPN (dark and light green) domains (Garcia-Doval et al. 2020). (B) T. onnurineus (Ton) Csm6 dimer in complex with its cyclic tetra-adenylate (cA4) activator. TonCsm6 is a nonspecific ribonuclease consisting of CARF and HEPN domains (Jia et al. 2019c). (C) S. islandicus (Sis) Csx1 hexamer in complex with its cA4 activator. SisCsx1 dimers form a hexamer upon cA4 binding and RNA is cleaved at three distinct active sites within the interior of the hexamer (Molina et al. 2019). (D) S. solfataricus (Sso) Csa3 dimer. SsoCsa3 is a cA4 stimulated transcription regulator consisting of CARF and a helix-turn-helix DNA binding domain (yellow and orange) (Lintner et al. 2011). (E) T. thermophilus (Tth) CRISPR ancillary nuclease 1 (Can1) monomer in complex with its cA4 activator. TthCan1 nicks super-coiled DNA and is comprised of two CARF domains and a PD-D/ExK family nuclease domain (salmon colored) (McMahon et al. 2020). (F) T. succinifaciens cyclic oligoadenylate activated RNase and DNase 1 (Card1)/Can2 dimer in complex with its cA4 activator. Card1/Can2 is related to Can1 and is a dual-specificity nuclease with CARF and PD-D/ExK nuclease domains (red and salmon) (Rostøl et al. 2021; Zhu et al. 2021). (G) E. coli (Eco) NucC hexamer in complex with its cyclic tri-adenylate (cA3) activator. EcoNucC trimers assemble into a hexamer upon cA3 binding and degrades dsDNA (Lau et al. 2020). NucC is related to restriction endonucleases and binds cA3 at a protein domain unrelated to the CARF family.