FIGURE 1.
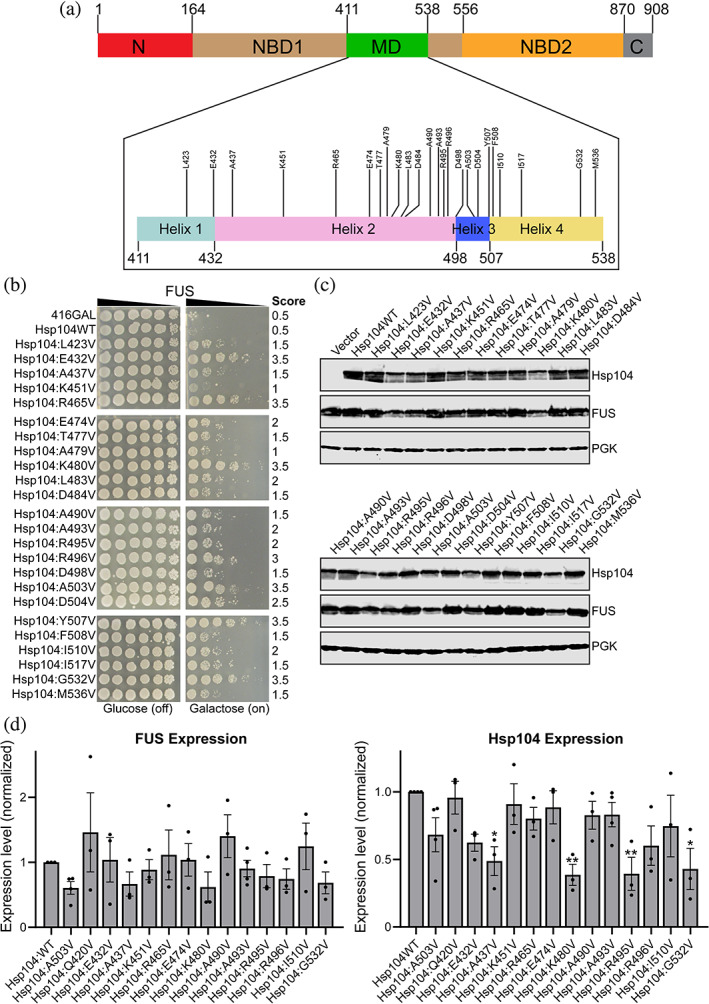
Scanning mutagenesis of the Hsp104 middle domain reveals Hsp104 variants that suppress FUS toxicity. (a) Domain map of Hsp104 shows the location of the MD (green) and its four helices (inset). Variants that suppress FUS toxicity are shown in the inset of the MD. (b) w303aΔhsp104‐pAG‐303GAL‐FUS yeast were transformed with Hsp104 variants or vector. Strains were serially diluted five‐fold and spotted in duplicate onto glucose (non‐inducing, left) or galactose (inducing, right) media. Scores are based on number of spots from an average of three replicates. Only variants that suppress FUS toxicity are shown, see Figure S1 for the full set of spotting assays. (c) Strains in (b) were induced for 5 hr, lysed, and immunoblotted for Hsp104, FUS, and 3‐phosphogylcerate kinase (PGK; loading control). (d) Quantification of FUS (left) and Hsp104 (right) immunoblots for selected strains. Values were normalized to the 3‐PGK loading control and are expressed relative to Hsp104WT. Expression of FUS and Hsp104 levels were compared to the strain expressing FUS and Hsp104WT using a one‐way ANOVA with a Dunnett's multiple comparisons test (N ≥ 3, individual data points shown as dots, bars show mean ± SEM, all p values >.05 except where denoted: *p < .05, **p < .01)
