FIGURE 1.
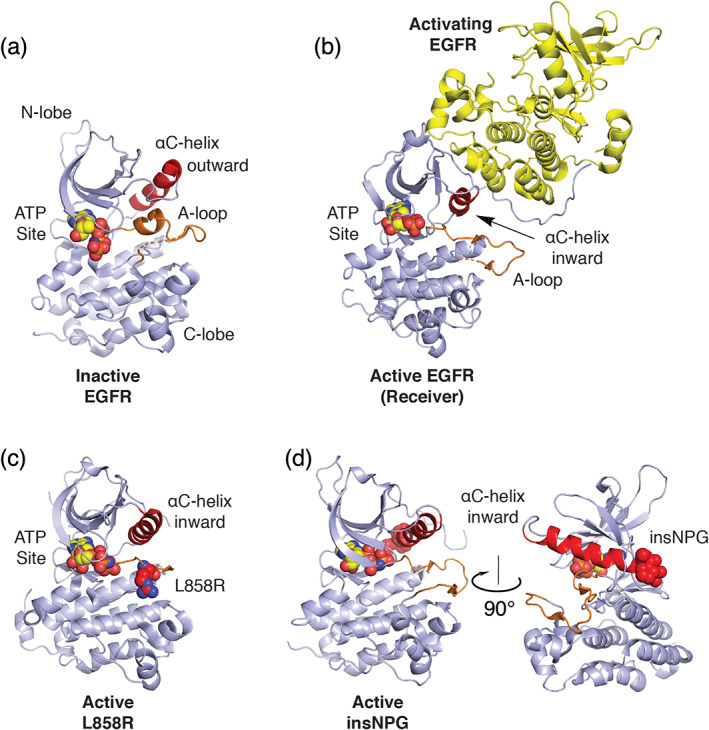
Structural basis for normal and oncogenic EGFR activation. (a) Structure of the EGFR kinase in its inactive state (PDB ID 2GS7). 28 A helical segment in the activation loop (A‐loop, orange) locks the αC‐helix (red) in an outward conformation. (b) Structure of the active kinase (receiver) promoted through asymmetric dimerization with an activating kinase domain. Based on PDB ID 2GS6. 28 (c) Structure of the EGFR kinase containing the oncogenic L858R activating mutation. The mutant Arg858 side chain is shown in space fill with carbon atoms in red (PDB ID 2ITV). 29 Residue L858 lies in the helical portion of the activation segment in the inactive state and the resulting Arginine (shown as red spheres) induces activation by destabilizing this element to allow the inward active position of the αC‐helix. (d) exon20 insNPG adds three amino acids (shown in red spheres) to the base of the αC‐helix sterically reinforcing the kinase in the active conformation (PDB ID 4LRM). 30 In all structures, ATP analogues that define the substrate site are shown in spacefill
