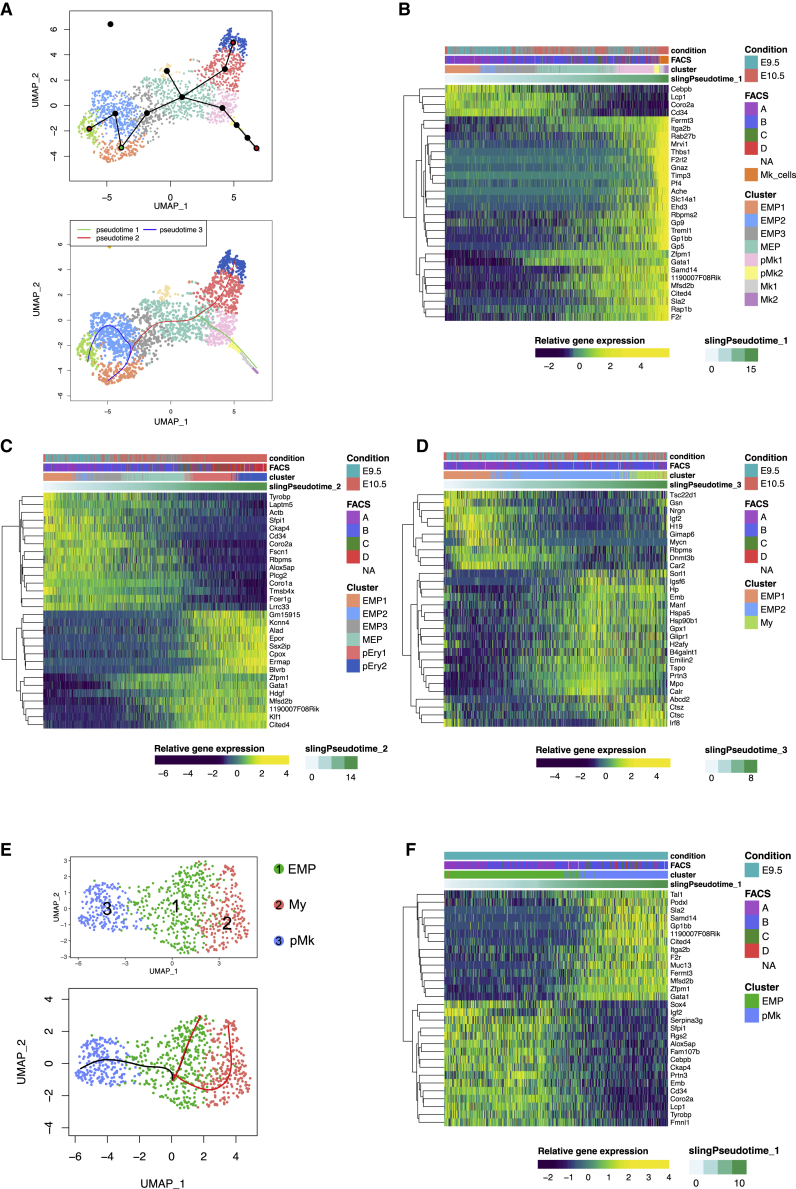
Figure 4.
Lineage trajectories of EMP commitment and differentiation in the yolk sac
(A) (Upper panel) Green node represents the starting point (EMP1) while red nodes represent the finishing points indicated to Slingshot to create the lineage trajectories. (Lower panel) Three lineage trajectories inferred by Slingshot toward Mk2 (green), pEry2 (red), and My clusters (blue).
(B) Heatmap of key genes of the trajectory from EMP1 to Mk2. Heatmap representing the differentially expressed genes along the trajectory.
(C) Heatmap of key genes of the trajectory from EMP1 toward Erythroid progenitors (pEry2).
(D) Heatmap of key genes of the trajectory from EMP1 to Myeloid progenitor cluster (My).
(E) Upper panel, UMAP representation of YS E9.5 Csf1ricreRosayfp YFP+ Kit+ cells colored by cluster: EMP (green), Myeloid (red) and Mk progenitor (blue) cluster. Lower panel, Slingshot infers two trajectories starting from EMP toward pMk or My. See also Figure S4.
(F) Heatmap of key genes of the trajectory of E9.5 EMP to Mk progenitor cluster (pMk). Top 30 genes (30 smallest p values) for the trajectories are represented in all the heatmaps. Please also see Figure S4.