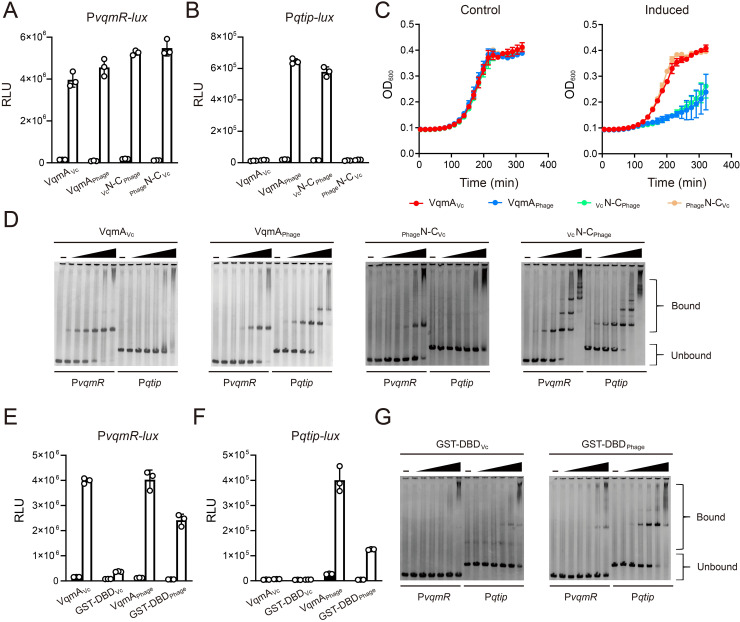
Fig 1. Promoter DNA-binding selectivity is conferred by the VqmA DBD.
(A and B) Normalized reporter activity from Δtdh E. coli harboring (A) PvqmR-lux or (B) Pqtip-lux and arabinose-inducible VqmAVc, VqmAPhage, VcN-CPhage, or PhageN-CVc. Black, no arabinose; white, 0.2% arabinose. Data are represented as mean ± SD (error bars) with n = 3 biological replicates. (C) Growth curves of the Δtdh ΔvqmAVc V. cholerae harboring phage VP882 vqmAPhage::Tn5 and arabinose-inducible VqmAVc, VqmAPhage, VcN-CPhage, or PhageN-CVc in medium lacking (Control) or containing 0.2% arabinose (Induced). (D) EMSAs showing binding of VqmA proteins to PvqmR and Pqtip DNA. From left to right are, VqmAVc, VqmAPhage, PhageN-CVc, and VcN-CPhage. 25 nM PvqmR or Pqtip DNA was used in all EMSAs with no protein (designated -) or 2-fold serial dilutions of proteins. The lowest and highest protein (dimer) concentrations are 18.75 nM and 600 nM, respectively. (E and F) Normalized reporter activity from WT E. coli as in panels A and B harboring arabinose-inducible VqmAVc, GST-DBDVc, VqmAPhage, and GST-DBDPhage. (G) EMSAs showing binding of GST-DBDVc and GST-DBDPhage to PvqmR and Pqtip DNA. Probe and protein concentrations as in panel D.