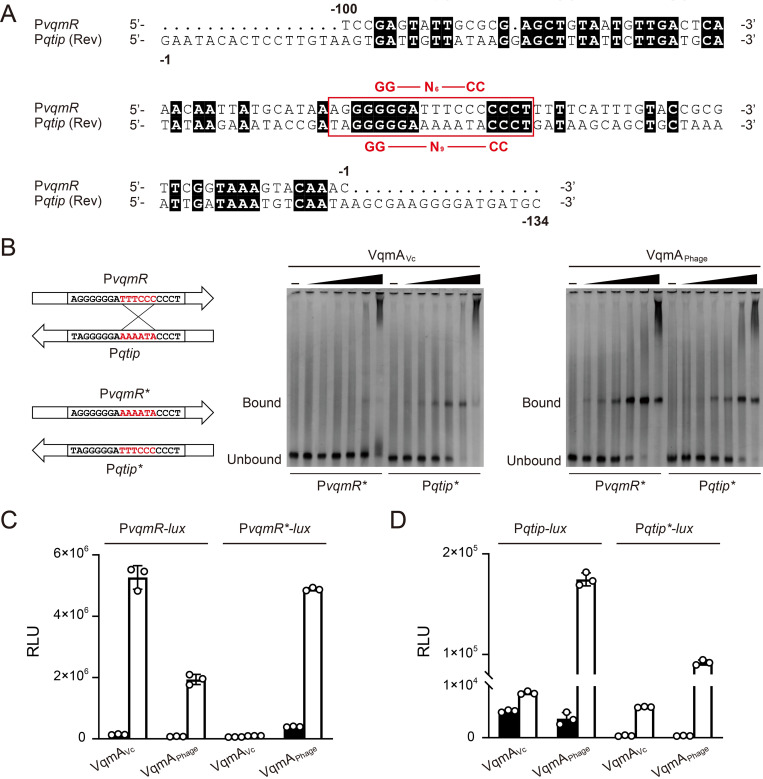
Fig 2. Promoter selectivity is reversed by exchanging key nucleotide fragments.
(A) DNA sequence alignment (ClustalW) of PvqmR and Pqtip. The reverse strand of Pqtip is shown. Numbering indicates positions relative to the transcription start sites. Identical nucleotides are designated with black shading. The reported 18-bp DNA stretch in PvqmR required for VqmAVc to bind (2,11) and the corresponding region in Pqtip are highlighted in the red box. The GG-N6-CC palindrome in PvqmR (2,11) and the recently identified GG-N9-CC palindrome in Pqtip (15) are indicated above and below the red box, respectively. (B) EMSAs showing binding of the designated VqmA proteins to PvqmR* and Pqtip* DNA. The cartoon at the left illustrates the key sequences exchanged in the probes. Probe and protein concentrations as in Fig 1D. (C) Normalized reporter activity from Δtdh E. coli harboring PvqmR-lux or PvqmR*-lux and arabinose-inducible VqmAVc or VqmAPhage. Black, no arabinose; white, 0.2% arabinose. Data are represented as mean ± SD (error bars) with n = 3 biological replicates. (D) As in C for Pqtip-lux or Pqtip*-lux.