Fig. 3. Neanderthal ancestry inferred in modern humans.
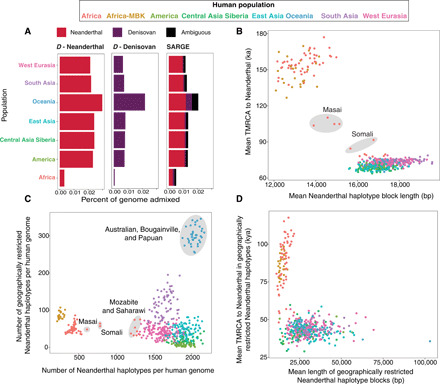
(A) Genome-wide percent Neanderthal, Denisovan, and ambiguous (either Neanderthal or Denisovan) across SGDP populations, using the ARG and an estimator based on the D statistic. D statistic calculations considered only one archaic population at a time as introgressor and thus do not detect ambiguous ancestry and also might count some Denisovan ancestry as Neanderthal and vice versa. (B) For individual phased human genome haplotypes (points), mean TMRCA with Neanderthal in Neanderthal haplotype blocks (y axis) and mean Neanderthal haplotype block length (x axis) for all Neanderthal-introgressed haplotypes. TMRCA calculations assume a total of 6.5-Ma human-chimpanzee divergence and branch shortening values from (1), with a mutation rate of 1 × 10−9 per site per year. bp, base pair. (C) Overall number Neanderthal haplotype blocks versus geographically restricted (unique to a 3000-km radius) Neanderthal haplotype blocks. (D) Same as (B), but limited to geographically restricted (unique to a 3000-km radius) Neanderthal haplotype blocks. Only haplotypes with more than 10 geographically restricted segments are shown.
