Fig. 7. Colonization of GF NOD with bacteria alters immune cell cytokine secretion and function.
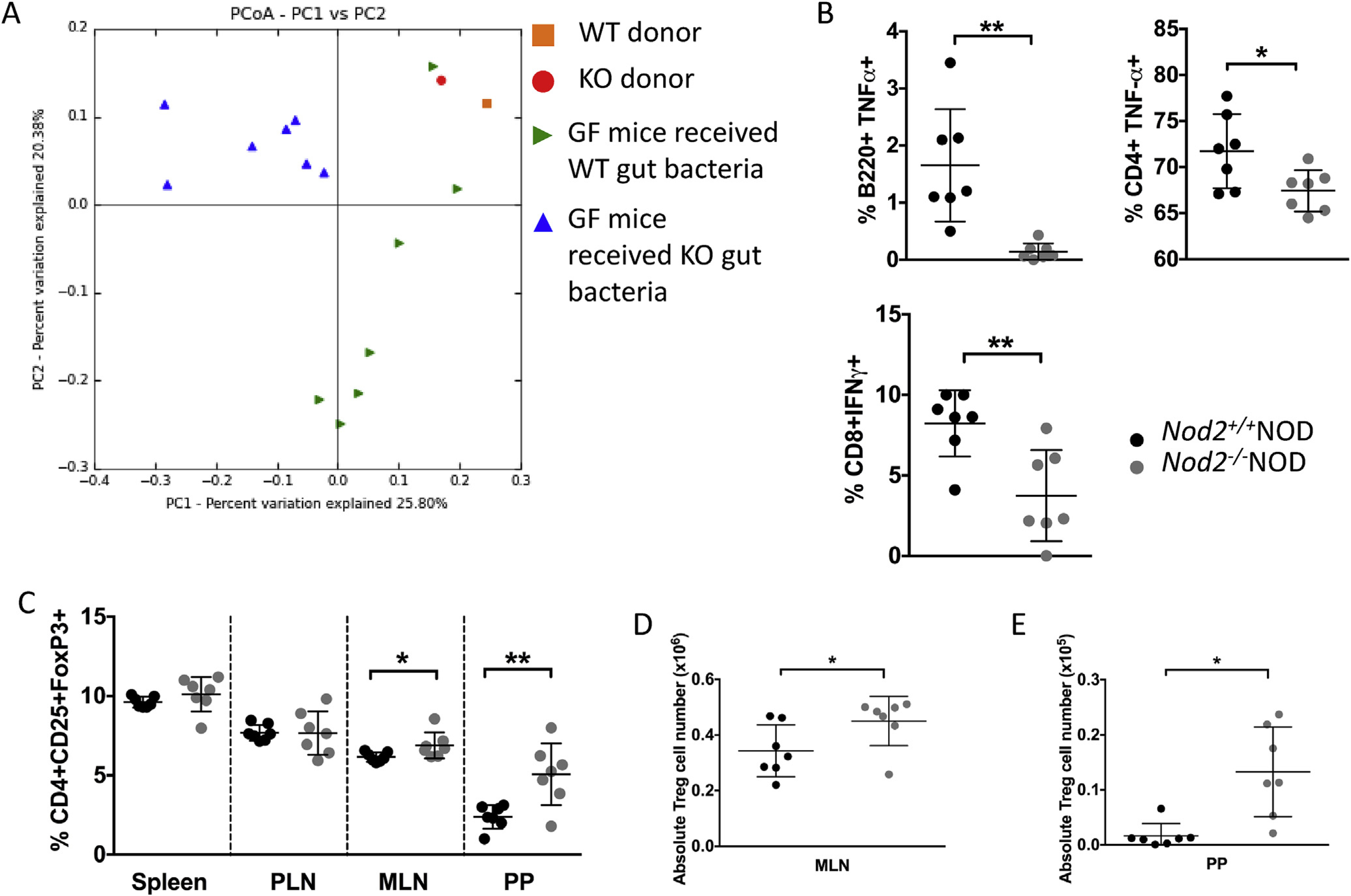
GF NOD mice were gavaged once with 2 × 108 CFU fecal bacteria from 4 to 5 week old Nod2−/−NOD KO or Nod2+/+NOD WT mice. Bacterial DNA was extracted from fecal pellets from WT or KO gavaged ex-GF NOD mice (n = 7/group) and the original bacteria donors. Pyrosequencing was conducted using the Ion Torrent PGM sequencing system. The sequencing data were analyzed with QIIME software package and UPARSE pipeline to pick operational taxonomic units (OTUs). Taxonomy assignment was performed at phylum level using representative sequences of each OTU. Principal component analysis (unweighted) of beta diversity of the gut microbiota from WT gavaged and KO gavaged mice is shown (A). 2–3 weeks later mice were sacrificed for immunophenotyping. Intracellular staining was conducted on B-cells (gated on live single TCRbeta-CD11b−CD11c−B220+) and T-cells (gated on live single TCRbeta+B22− cells then CD4 or CD8) from the pancreatic lymph nodes (B). FoxP3 staining was conducted following the instructions from the eBioscience staining kit for cells from spleen, pancreatic lymph nodes (PLN), mesenteric lymph nodes (MLN) and Peyer’s patches (PP). Cells were gated on live single B220−TCRbeta+CD8a−CD4+CD25+FoxP3+ cells (C). Absolute numbers were calculated for MLN (D) and PP (E) Treg cells. All data shown are pooled from two independent experiments (n = 3–4/experiment). Statistical analysis was performed using Student’s t-test, *P < 0.05 or **P < 0.0001. Lines represent the mean and the standard deviation.
