Figure 1: Oncogenic RET fusions.
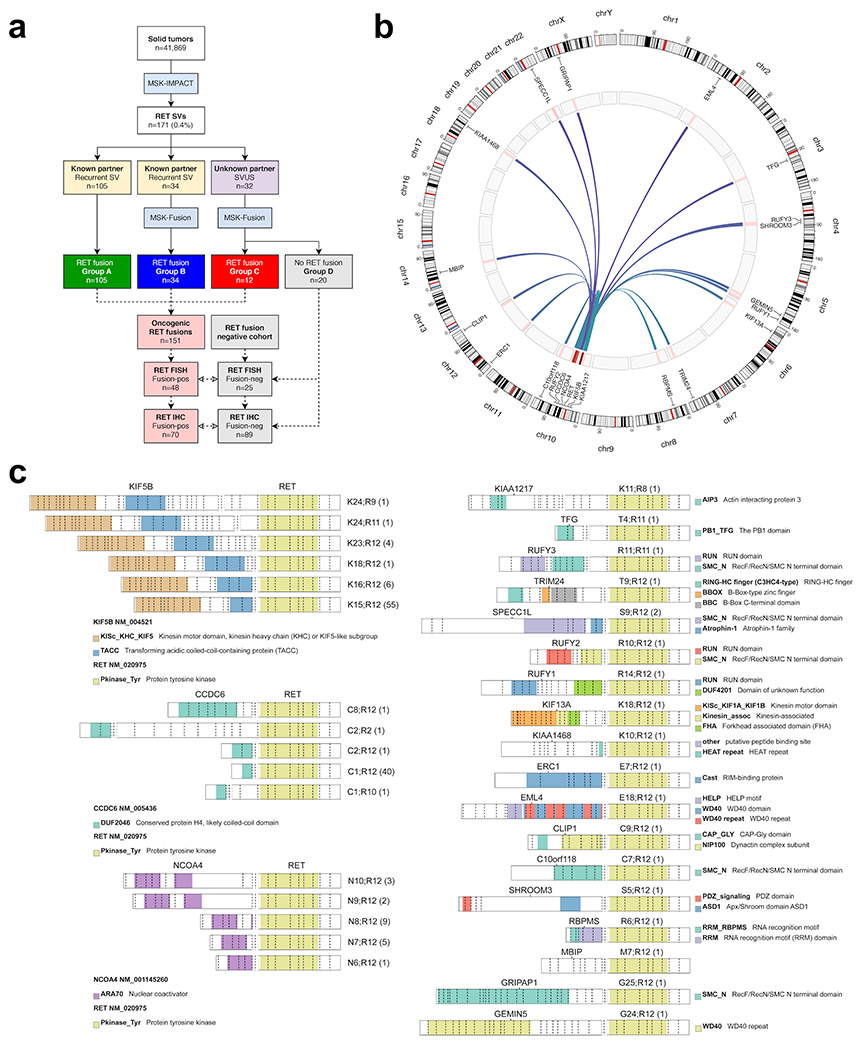
(A) Solid tumors were screened for RET fusions using a targeted DNA-based NGS assay (MSK-IMPACT). A subset of recurrent oncogenic RET structural variants (SVs) and all RET structural variants of unknown significance (SVUS) were confirmed using a targeted RNA-based NGS panel (MSK-Fusion). Through this approach, a total of 151 oncogenic RET fusions were identified. A subset of these fusions was further tested by break-apart FISH and immunohistochemistry (IHC). (B) Distribution of oncogenic RET fusions (n=151) across the genome. Red bars indicate frequency for each fusion type. (C) Distribution of RET fusions involving KIF5B, CCDC6, and NCOA4 partners (left panel) and other rare fusion partners (right panel). The exonic breakpoints and the observed frequency (in parenthesis) are provided for each fusion. The colors correspond to different protein domains.
